Hummer 2016 Biochim Biophys Acta
| Hummer G, Wikström M (2016) Molecular simulation and modeling of complex I. Biochim Biophys Acta 1857:915-21. https://doi.org/10.1016/j.bbabio.2016.01.005 |
Hummer G, Wikstroem Marten KF (2016) Biochim Biophys Acta
Abstract: Molecular modeling and molecular dynamics simulations play an important role in the functional characterization of complex I. With its large size and complicated function, linking quinone reduction to proton pumping across a membrane, complex I poses unique modeling challenges. Nonetheless, simulations have already helped in the identification of possible proton transfer pathways. Simulations have also shed light on the coupling between electron and proton transfer, thus pointing the way in the search for the mechanistic principles underlying the proton pump. In addition to reviewing what has already been achieved in complex I modeling, we aim here to identify pressing issues and to provide guidance for future research to harness the power of modeling in the functional characterization of complex I. This article is part of a Special Issue entitled Respiratory complex I, edited by Volker Zickermann and Ulrich Brandt.
• Bioblast editor: Gnaiger E
Labels:
Preparation: Enzyme
Enzyme: Complex I
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
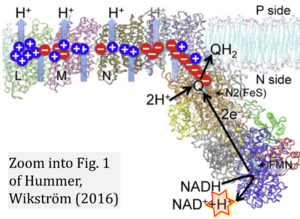
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.