Description
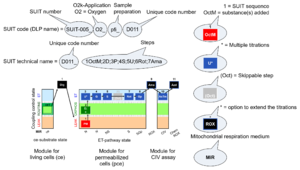
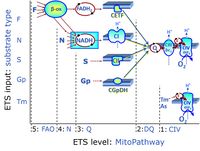
The SUIT protocol name starts with (i) the SUIT category which shows the ETS substrate types applied (controlling ETS pathway types; e.g. S, NS, FNS, FNSGp), independent of the actual sequence of titrations. (ii) A further distinction is provided in the SUIT name by listing in parentheses the N-type substrates applied, again independent of the sequence of titrations, e.g. NS(GM), NS(PM), FNSGp(PGM). (iii) A sequentially selected number is added, e.g. SUIT_FNS(PM)01 (see Coupling-pathway control diagram).
The SUIT sequence starts with the category of the SUIT protocol, and separates the category with an underline dash from the sequence of titration steps (mark names, #X, separated by a space). The Marks define the section of a respiratory state in the SUIT protocol. The Mark name contains the sequential number and the metabolic control variable, X. The metabolic control variable is the name of the preceding SUIT event. The list of MitoPedia SUIT protocols can thus be sorted by the SUIT protocol category (sorting by SUIT protocol name), and by SUIT protocol subcategory which is listed as the abbreviation of the SUIT protocol name. The SUIT protocol pattern is best illustrated by a coupling-pathway control diagram.
Abbreviation: SUITp-Names
Reference: MiPNet21.06 SUIT RP
MitoPedia concepts:
MiP concept,
SUIT concept
Contributed by Gnaiger Erich 2016-02-01; edited 2016-09-05.