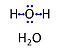Semantic search
| Term | Abbreviation | Description |
|---|---|---|
| Q-pools | Q | Different Q-pools are more or less clearly distinguished in the cell, related to a variety of models describing degress of Q-pool behavior. (1) CoQ-pools are distinguished according to their compartmentation in the cell: mitochondrial CoQ (mtCoQ) and CoQ in other organelles versus plasma-membrane CoQ. (2) The total mitochondrial CoQ-pool mtCoQ is partitioned into an ETS-reactive Q-pool, Qra, and an inactive mtCoQ-pool, Qia. (2a) The Qra-pool is fully reduced in the form of quinol QH2 under anoxia, and fully oxidized in the form of quinone in aerobic mitochondrial preparations incubated without CHNO-fuel substrates. Intermediate redox states of Qra are sensitive to pathway control and coupling control of mitochondrial electron transfer and OXPHOS. (2b) The Qia-pool remains partially reduced and oxidized independent of aerobic-anoxic transitions. The redox state of Qia is insensitive to changes in mitochondrial respiratory states. (3) The Qra-pool is partitioned into Q with Q-pool behavior according to the fluid-state model (synonymous: random-collision model) and Q tightly bound to supercomplexes according to the solid-state model. The two models describe the extremes in a continuum of homogenous or heterogenous Q-pool behavior. The CII-Q-CIII segment of the S-pathway is frequently considered to follow homogenous Q-pool behavior participating in the Qhom-pool, whereas the CI-Q-CIII segment of the N-pathway indicates supercomplex organization and metabolic channeling with different degrees of Q-pool heterogeneity contributing to the Qhet-pool. |
| Quantity | Q | A quantity is the attribute of a phenomenon, body or substance that may be distinguished qualitatively and determined quantitatively. A dimensional quantity is a number (variable, parameter, or constant) connected to its dimension, which is different from 1. {Quote} The value of a quantity is generally expressed as the product of a number and a unit. The unit is simply a particular example of the quantity concerned which is used as a reference, and the number is the ratio of the value of the quantity to the unit. {end of Quote: Bureau International des Poids et Mesures 2019 The International System of Units (SI), p. 127)}. |
| R-L control efficiency | jR-L | |
| R-L net ROUTINE capacity | R-L | |
| R/E control ratio | R/E | |
| ROUTINE respiration | R | |
| RT | RT versus RT | RT indicates room temperature or 25 °C. RT is the gas constant R [kJ/mol] multiplied by absolute temperature T [K]. This is the motive force quantum in the amount format (Gnaiger 2020 BEC MitoPathways). |
| Raw signal of the oxygen sensor | R | The raw signal of the polarographic oxygen sensor is the current Iel [µA], 1 µA = 10-6 C·s-1, (DatLab 8) or the electric potential difference (voltage) [V], 1 V = 1 J·C-1, obtained after a current-to-voltage conversion in the O2k (DatLab 7 and previous versions). |
| Reactive nitrogen species | RNS | Reactive nitrogen species, RNS, are nitric oxide-derived oxidants. The main source of RNS is nitric oxide (NO•). NO• plays an important role in cell signaling and in oxidative-nitrosative stress. |
| Reactive oxygen species | ROS | Reactive oxygen species, ROS, are molecules derived from molecular oxygen, including free oxygen radicals, which are more reactive than O2. Physiologically and pathologically important ROS include superoxide, the hydroxyl radical and hydroxide ion, hydrogen peroxide and other peroxides. These are important in cell signalling, oxidative defence mechanisms and oxidative stress. |
| Reference material | RM | Reference material (RM) is material or substance one or more of whose property values are sufficiently homogeneous and well established to be used for the calibration of an apparatus, the assessment of a measurement procedure, or for assigning values to materials (adapted from VIM: 1993, 6.13). The adjective 'homogeneous' refers to the physical homogeneity between macroscopic parts of the material, not to any microheterogeneity between molecules of the analyte.Primary reference material is reference material having the highest metrological qualities and whose value is determined by means of a primary reference measurement procedure. The concept "primary calibrator" is subordinate to "calibrator" (see 3.7) and to "primary reference material". |
| Reference state | Z | The reference state Z (reference rate ZX) is the respiratory state with high flux in relation to the background state Y with low background flux YX. The transition between the background state and the reference state is a step brought about by a metabolic control variable X. If X stimulates flux (ADP, fuel substrate), it is present in the reference state but absent in the background state. If X is an inhibitor of flux, it is absent in the reference state but present in the background state. The reference state is specific for a single step to define the flux control efficiency. In contrast, in a sequence of multiple steps, the common reference state is frequently taken as the state with the highest flux in the entire sequence, as used in the definition of the flux control ratio. |
| References in BEC https-format | BEC https-format | References in BEC https-format show (1) the list of authors, (2) the year of publication in parentheses, (3) the title of the publication, and (4) the https://doi.org/ link. Removing all the unnecessary detail of journal name and pages, the focus is on authors, year, and title of the reference, which is a concept in line with DORA. The https-link then does the full job. In exceptional cases when there is no such link, the following formats would apply: a https://pubmed.ncbi.nlm.nih.gov/ link, a link directly to an Open Access pdf, or the old conventional format for the reference. Scientific journals apply yesterday's concepts in the various formats of references with bewildering abbreviations of journals, volumes, issues, page numbers. We can do today's job much better using the BEC https-format:
Compare with a conventional reference format in: Gnaiger E (2021) Beyond counting papers – a mission and vision for scientific publication. https://doi.org/10.26124/bec:2021-0005 |
| Repetitions | n | Repetitions of an experiment or assay are designed to obtain statistical information on the methodological precision of the measurements. A number of repetitions, n, of measurements are performed on the same sample, applying an identical experimental protocol to subsamples, without providing any information on variability between samples. |
| Replica | N | Replica are designed in scientific studies to evaluate the effect of uncontrolled variability on a result obtained from an experiment on a single sample, to describe the variability and distribution of experimental results, and to obtain statistical information such as the median or average for a defined sample size. It may be useful to make a terminological distinction between replica of experiments, N, designed to obtain statistical information on the population, and repetitions of experiments or assays, n, designed to obtain statistical information on the methodological precision of the measurements. The terms study, experiment and assay have to be defined carefully in this context. |
| Residual endogenous substrates | REN, Ren |
Ren may be higher than Rox. Correspondingly, Q and NAD are not fully oxidized in the REN state compared to the ROX state. In previous editions (including Gnaiger 2020 BEC MitoPathways), the REN state was not distinguished from the ROX state. However, in novel applications (Q-Module and NADH-Module), a distinction of these states is necessary. Care must be taken when assuming Ren as a substitute of Rox correction of mitochondrial respiration. |
| Residual oxygen consumption | ROX, Rox |
In previous editions, (including Gnaiger 2020 BEC MitoPathways), the REN state was not distinguished from the ROX state. However, in novel applications (Q-Module and NADH-Module), a distinction of these states is necessary. Care must be taken when assuming Ren as a substitute of Rox correction of mitochondrial respiration. |
| Resorufin | Res | Resorufin is a fluorescence probe used in various biological assays. Among others, it is the product obtained in the Horseradish peroxidase-catalyzed assay using Amplex Red for the measurement of H2O2 production. |
| Respiratory acceptor control ratio | RCR | The respiratory acceptor control ratio (RCR) is defined as State 3/State 4 [1]. If State 3 is measured at saturating [ADP], RCR is the inverse of the OXPHOS control ratio, L/P (when State 3 is equivalent to the OXPHOS state, P). RCR is directly but non-linearly related to the P-L control efficiency, jP-L = 1-L/P, with boundaries from 0.0 to 1.0. In contrast, RCR ranges from 1.0 to infinity, which needs to be considered when performing statistical analyses. In living cells, the term RCR has been used for the ratio State 3u/State 4o, i.e. for the inverse L/E ratio [2,3]. Then for conceptual and statistical reasons, RCR should be replaced by the E-L coupling efficiency, 1-L/E [4]. |
| Respiratory chain | RC | The mitochondrial respiratory chain (RC) consists of enzyme complexes arranged to form a metabolic system of convergent pathways for oxidative phosphorylation. In a general sense, the RC includes (1) the electron transfer pathway (ET-pathway), with transporters for the exchange of reduced substrates across the inner mitochondrial membrane, enzymes in the matrix space (particularly dehydrogenases of the tricarboxylic acid cycle), inner membrane-bound electron transfer complexes, and (2) the inner membrane-bound enzymes of the phosphorylation system. |
| Respiratory complexes | CI, CII, CIII, CIV, CETF, CGpDH, .. | Respiratory Complexes are membrane-bound enzymes consisting of several subunits which are involved in energy transduction of the respiratory system. » MiPNet article |
| Resting metabolic rate | RMR | Resting respiration or resting metabolic rate (RMR) is measured under standard conditions of an 8–12-h fast and a 12-h abstinence from exercise. In an exemplary study (Haugen 2003 Am J Clin Nutr), "subjects rested quietly in the supine position in an isolated room with the temperature controlled to 21–24° C. RMR was measured for 15–20 min. Criteria for a valid RMR was a minimum of 15 min of steady state, determined as a <10% fluctuation in oxygen consumption and <5% fluctuation in respiratory quotient". The main difference between RMR and BMR (basal metabolic rate) is the position of the subject during measurement. Resting metabolic rate is the largest component of the daily energy budget in most human societies and increases with physical training state (Speakman 2003 Proc Nutr Soc). |
| Reverse electron flow from CII to CI | RET | Reverse electron flow from CII to CI stimulates production of ROS when mitochondria are incubated with succinate without rotenone in the LEAK state at a high mt-membrane potential. Depolarisation of the mt-membrane potential (e.g. after ADP addition to stimulate OXPHOS) leads to inhibition of RET and therefore, decrease of RET-initiated ROS production. RET can be also measured when mitochondria are respiring using Gp without rotenone in the LEAK state. Addition of IQ-side inhibitors (ubiquinone-binding side of CI) of CI usually block RET. The following SUIT protocols allow you to measure RET-initiated H2O2 flux in mitochondrial preparations: SUIT-009 and SUIT-026. |
| Rhodamine 123 | Rh123 | Rhodamine 123 (Rh123) is an extrinsic fluorophore and can be used as a probe to determine changes in mitochondrial membrane potential. Rh123 is a lipophilic cation that is accumulated by mitochondria in proportion to Δψmt. Using ethanol as the solvent, the excitation maximum is 511 nm and the emission maximum is 534 nm. The recommended excitation and emission wavelengths in PBS are 488 and 515-575 nm, respectively (Sigma-Aldrich). |
| Rotenone | Rot | Rotenone is an inhibitor of Complex I (CI) and thus inhibits NADH oxidation. It inhibits the transfer of electrons from iron-sulfur clusters in CI to ubiquinone via binding to the ubiquinone binding site of CI. See also Succinate pathway. |
| Ruthenium red | RR | Ruthenium red (synonym: ammoniated ruthenium oxychloride) inhibits the mitochondrial Ca2+ uniporter. However, in addition it has been shown to interact with and inhibit a large number of other proteins, including ion channels particularly of the Transient Receptor Potential Vanilloid (TRPV) family [1], Ca2+-ATPases, and, importantly, the voltage-dependent anion channel (VDAC) [2]. |
| S/NS pathway control ratio | S/NS | The S/NS pathway control ratio is obtained when rotenone (Rot) is added to the NS-pathway control state in a defined coupling control state. The reversed protocol, adding N-type substrates to a S-pathway control state as the background state does not provide a valid estimation of S-linked respiration with succinate in the absence of Rot, since oxaloacetate accumulates as a potent inhibitor of succinate dehydrogenase (CII). |
| SF6847 | SF6847 | SF6847 (C18H22N2O), also known as tyrphostin A9 or malonoben, is a protonophore and a very potent uncoupler of oxidative phosphorylation, being used in the nM range. Like all uncouplers, SF6847 concentrations must be titrated carefully to evaluate the optimum concentration for maximum stimulation of mitochondrial respiration, particularly to avoid inhibition of respiration at higher concentrations. |
| SGp-pathway control state | SGp | SGp: Succinate & Glycerophosphate. MitoPathway control state: SGp; obtained with OctPGMSGp(Rot) SUIT protocol: SUIT-001 and ((SUIT-002 |
| STPD | STPD | At standard temperature and pressure dry (STPD: 0 °C = 273.15 K and 1 atm = 101.325 kPa = 760 mmHg), the molar volume of an ideal gas, Vm, and Vm,O2 is 22.414 and 22.392 L∙mol-1, respectively. Rounded to three decimal places, both values yield the conversion factor of 0.744 from units used in spiroergometry (VO2max [mL O2·min-1]) to SI units [µmol O2·s-1]. For comparison at normal temperature and pressure dry (NTPD: 20 °C), Vm,O2 is 24.038 L∙mol-1. Note that the SI standard pressure is 100 kPa, which corresponds to the standard molar volume of an ideal gas of 22.711 L∙mol-1 and 22.689 L∙mol-1 for O2. |
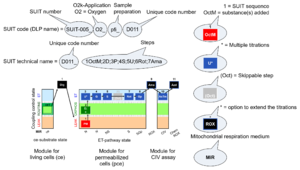
| SUIT | SUIT | SUIT is the abbreviation for Substrate-Uncoupler-Inhibitor Titration. SUIT protocols are used with mt-preparations to study respiratory control in a sequence of coupling and pathway control states induced by multiple titrations within a single experimental assay. These studies use biological samples economically to gain maximum information with a minimum amount of cells or tissue. |
| SUIT protocol library | SUITs | The Substrate-uncoupler-inhibitor titration (SUIT) protocol library contains a sequential list of SUIT protocols (D001, D002, ..) with links to the specific SUIT pages. Classes of SUIT protocols are explained with coupling and substrate control defined for mitochondrial preparations. |
| SUIT protocol names | SUITp-Names |
The SUIT protocol name starts with (i) the SUIT category which shows the Electron-transfer-pathway states (ET pathway types; e.g. N, S, NS, FNS, FNSGp), independent of the actual sequence of titrations. (ii) A further distinction is provided in the SUIT name by listing in parentheses the substrates applied in the N-pathway control states, again independent of the sequence of titrations, e.g. NS(GM), NS(PM), FNSGp(PGM). (iii) A sequentially selected number is added, e.g. SUIT_FNS(PM)01 (see Coupling/pathway control diagram). The systematic name of a SUIT protocol starts with the SUIT category, followed by an underline dash and the sequence of titration steps (mark names, #X, separated by a comma). The Marks define the section of a respiratory state in the SUIT protocol. The Mark name contains the sequential number and the metabolic control variable, X. The metabolic control variable is the name of the preceding SUIT event. The MitoPedia list of SUIT protocols can be sorted by the short name or the systematic name (hence by SUIT protocol category. The SUIT protocol pattern is best illustrated by a coupling/pathway control diagram. |
| SUIT protocol pattern | SUITp-Pattern | The SUIT protocol pattern describes the type of the sequence of coupling and substrate control steps in a SUIT protocol, which may be liner, orthogonal, or diametral. |
| SUIT reference protocol | SUIT RP | The substrate-uncoupler-inhibitor titration (SUIT) reference protocol, SUIT RP, provides a common baseline for comparison of mitochondrial respiratory control in a large variety of species, tissues and cell types, mt-preparations and laboratories, for establishing a database on comparative mitochondrial phyisology. The SUIT RP consists of two harmonized SUIT protocols (SUIT-001 - RP1 and SUIT-002 - RP2). These are coordinated such that they can be statistically evaluated as replicate measurements of cross-linked respiratory states, while additional information is obtained when the two protocols are conducted in parallel. Therefore, these harmonized SUIT protocols are complementary with their focus on specific respiratory coupling and pathway control aspects, extending previous strategies for respirometrc OXPHOS analysis.
|
| SUIT-001 | RP1 |  |
| SUIT-001 O2 ce-pce D003 | RP1 ce-pce |  |
| SUIT-001 O2 ce-pce D004 | RP1 ce-pce blood | 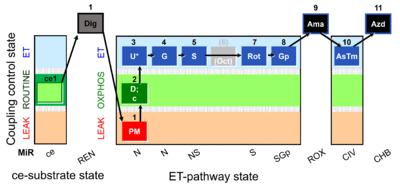 |
| SUIT-001 O2 mt D001 | RP1 mt | 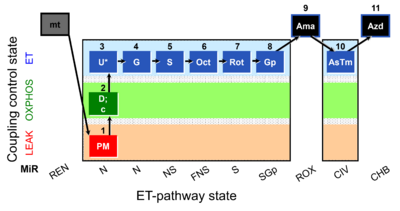 |
| SUIT-001 O2 pfi D002 | RP1 pfi | 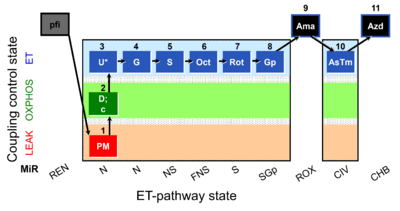 |
| SUIT-002 | RP2 | 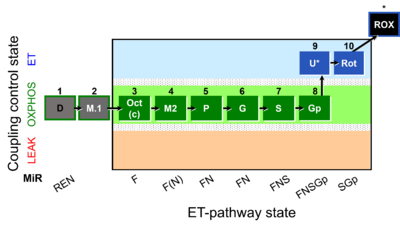 |
| SUIT-002 O2 ce-pce D007 | RP2 ce-pce | 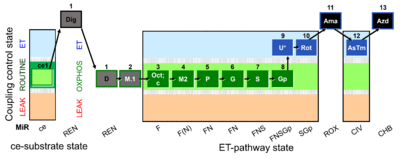 |
| SUIT-002 O2 ce-pce D007a | RP2 ce-pce blood |  |
| SUIT-002 O2 mt D005 | RP2 mt | 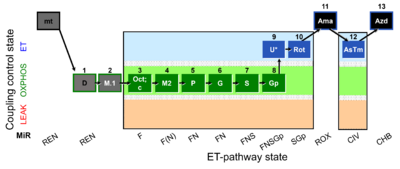 |
| SUIT-002 O2 pfi D006 | RP2 pfi | 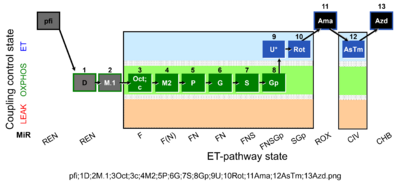 |
| SUIT-003 | CCP-ce | 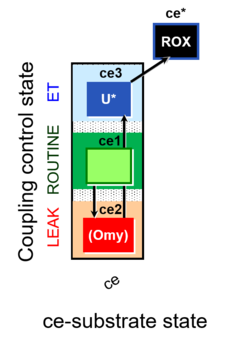 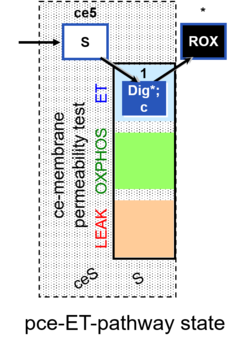 |
| SUIT-003 AmR ce D017 | CCP-ce S permeability test | 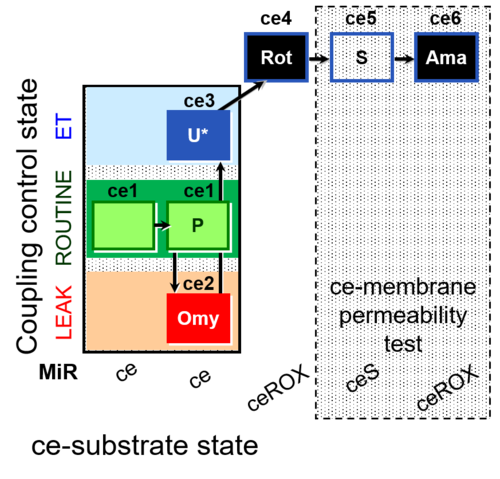 |
| SUIT-003 AmR ce D058 | AmR effect on ce | 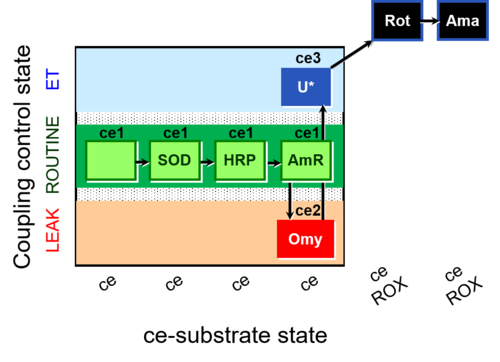 |
| SUIT-003 AmR ce D059 | AmR effect on ce - control | 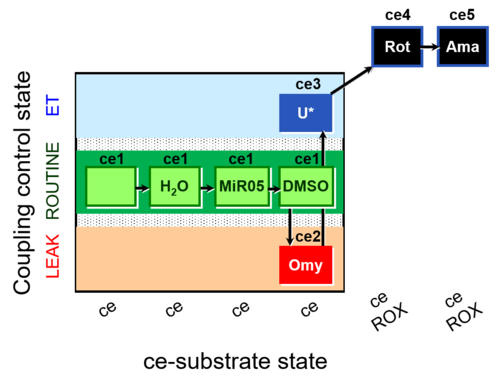 |
| SUIT-003 Ce1;ce1P;ce3U;ce4Glc;ce5M;ce6Rot;ce7S;1Dig;1c;2Ama;3AsTm;4Azd | cePMGlc,S | 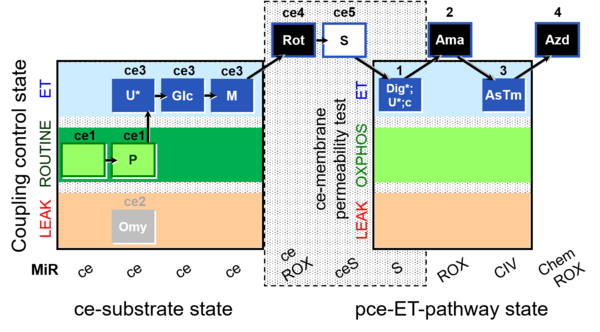 |
| SUIT-003 Ce1;ce1SD;ce2Omy;ce3U- | FNS(Oct,PGM) | 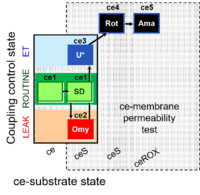 |
| SUIT-003 Ce1;ce1SD;ce3U;ce4Rot;ce5Ama | ceS | 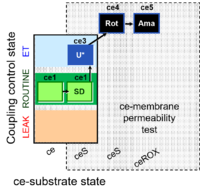 |
| SUIT-003 Ce1;ce2SD;ce3Omy;ce4U- | FNS(Oct,PGM) | 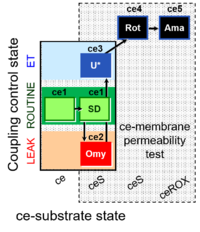 |
| SUIT-003 Ce1;ce2SD;ce3U;ce4Rot;ce5Ama | ceS |  |
| SUIT-003 Ce1;ce2U;ce3Rot;ce4S;ce5Ama | ceS |  |
| SUIT-003 Ce1;ce2U- | ce |  |
| SUIT-003 Ce1;ce3U;ce4Rot;ce5S;ce6Ama | ceS |  |
| SUIT-003 Ce1;ce3U- | ce | 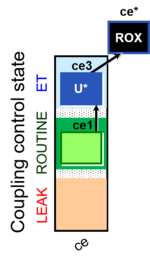 |
| SUIT-003 O2 ce D009 | CCP-ce short | 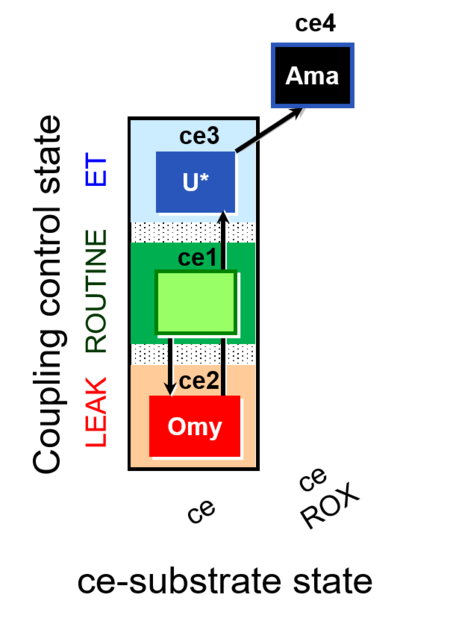 |
| SUIT-003 O2 ce D012 | CCP-ce(P) | 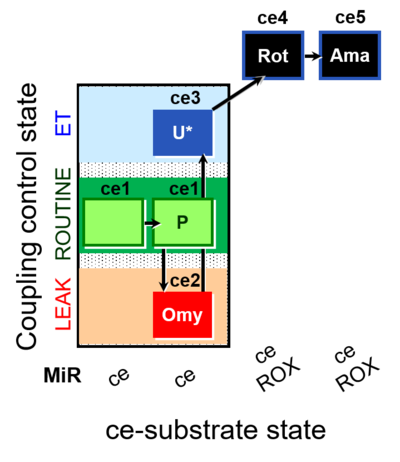 |
| SUIT-003 O2 ce D028 | CCP-ce S permeability test | 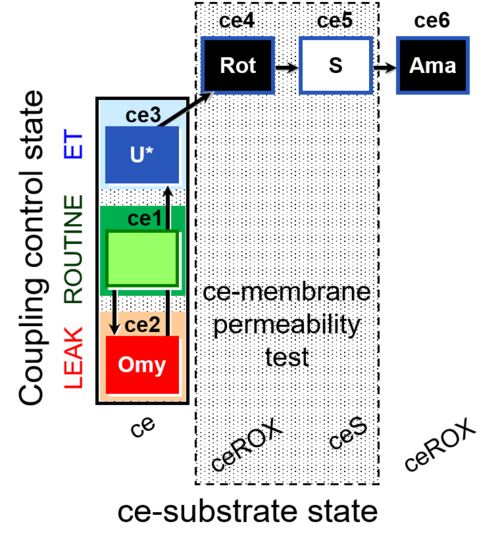 |
| SUIT-003 O2 ce D037 | CCP-ce Crabtree_R | 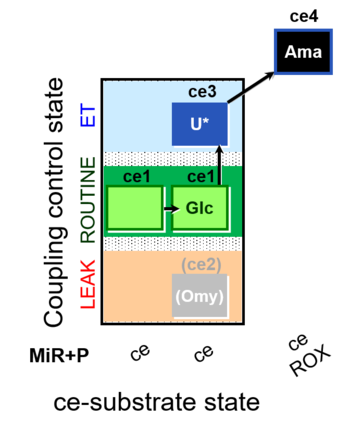 |
| SUIT-003 O2 ce D038 | CCP-ce Crabtree_E | 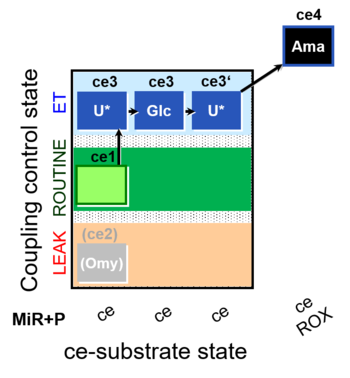 |
| SUIT-003 O2 ce D039 | CCP-ce microalgae | 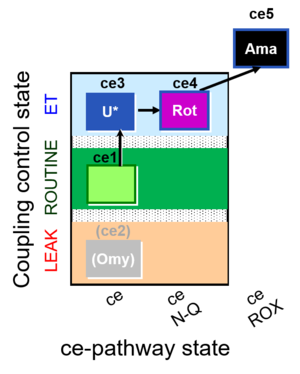 |
| SUIT-003 O2 ce D050 | CCP-ce Snv | 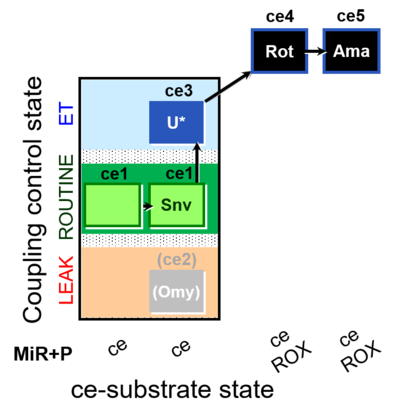 |
| SUIT-003 O2 ce D060 | CCP-ce Snv,Mnanv | 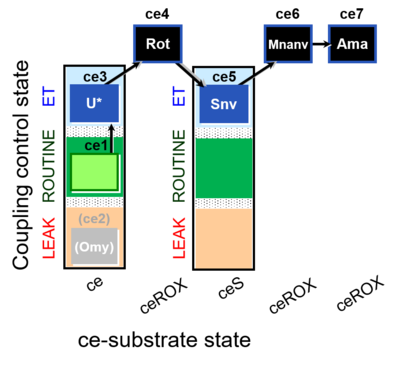 |
| SUIT-003 O2 ce D061 | CCP-ce Snv,Mnanv - control | 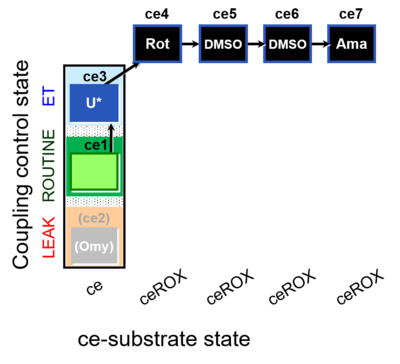 |
| SUIT-003 O2 ce D062 | CCP-ce Snv - control | 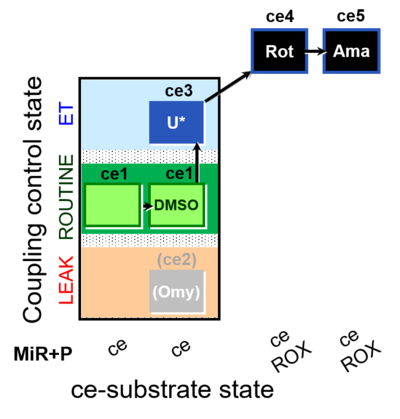 |
| SUIT-003 O2 ce-pce D013 | CCVP-Glc,M | 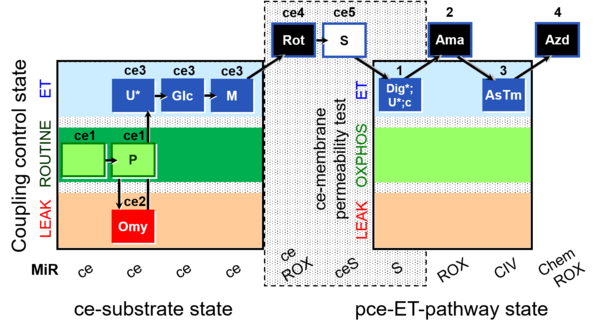 |
| SUIT-003 O2 ce-pce D018 | CCVP-Glc | 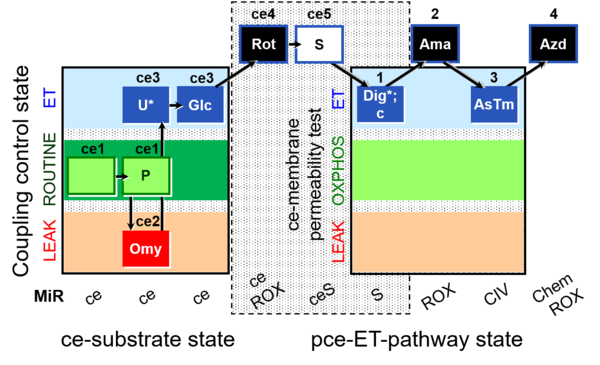 |
| SUIT-003 O2 ce-pce D020 | CCVP | 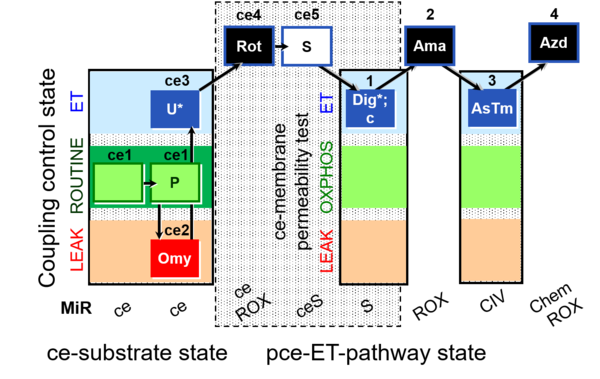 |
| SUIT-003 pH ce D067 | CCP-Crabtree with glycolysis inhibition | 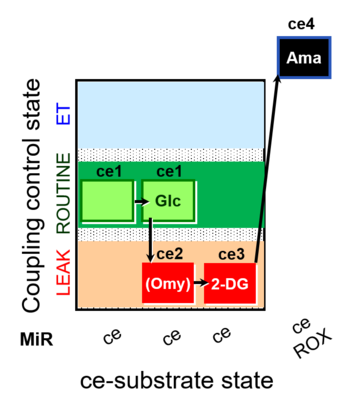 |
| SUIT-004 | RP1-short |  |
| SUIT-004 O2 pfi D010 | RP1-short pfi | 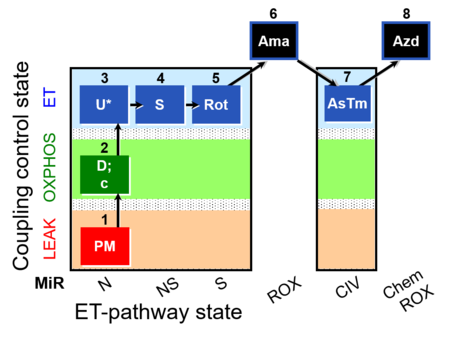 |
| SUIT-005 | RP2-short | 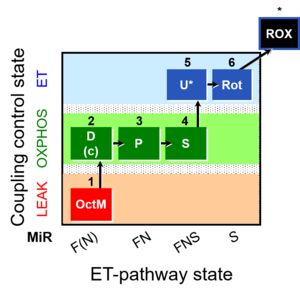 |
| SUIT-005 O2 pfi D011 | RP2-short pfi | 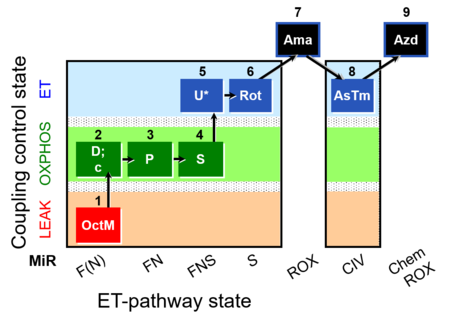 |
| SUIT-006 | CCP-mtprep | 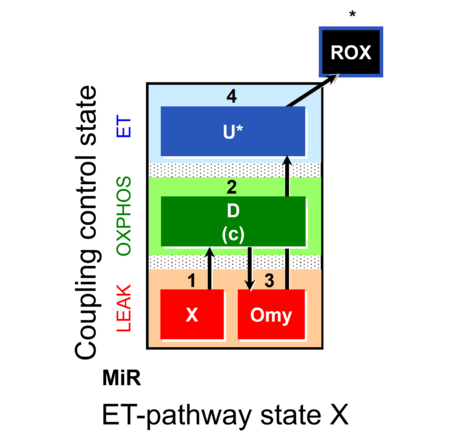 |
| SUIT-006 AmR mt D048 | CCP mt PM - AmR | 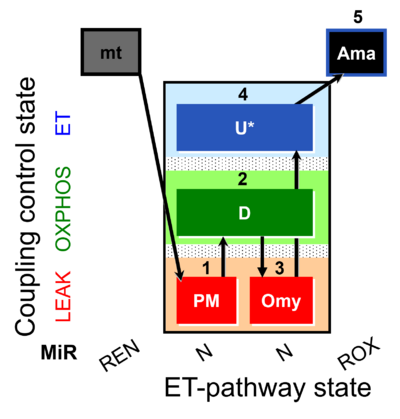 |
| SUIT-006 Fluo mt D034 | CCP mt PM - Fluo | 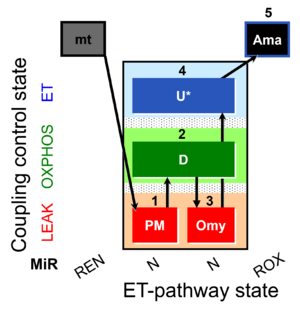 |
| SUIT-006 MgG ce-pce D085 | CCP MgG ce-pce | 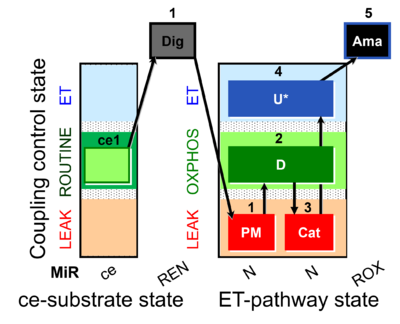 |
| SUIT-006 MgG mt D055 | CCP MgG mt | 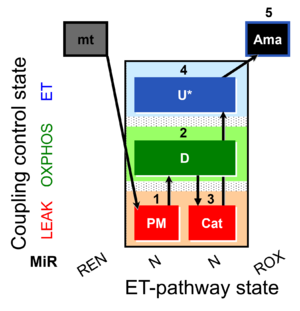 |
| SUIT-006 O2 ce-pce D029 | CCP ce-pce PM | 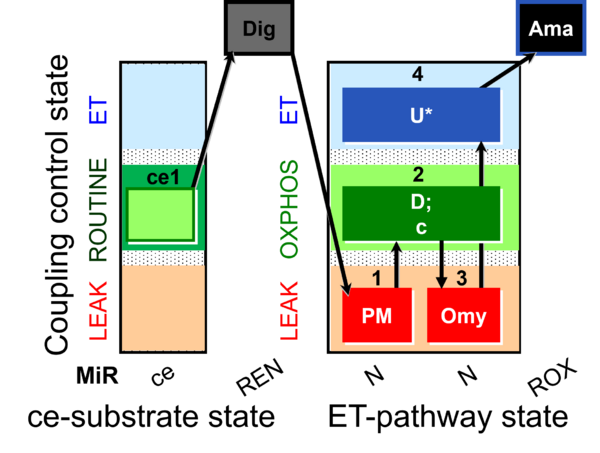 |
| SUIT-006 O2 mt D022 | CCP mt S(Rot) | 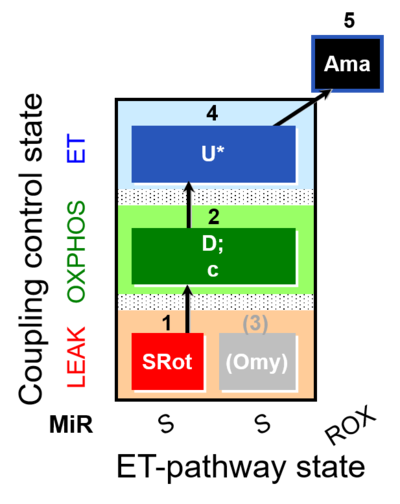 |
| SUIT-006 O2 mt D047 | CCP mt PM | 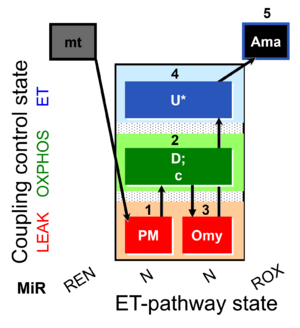 |
| SUIT-006 Q ce-pce D073 | CCP ce-pce S(Rot) | 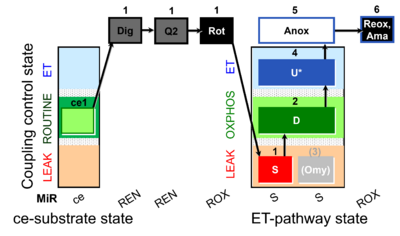 |
| SUIT-006 Q mt D071 | CCP mt S(Rot) | 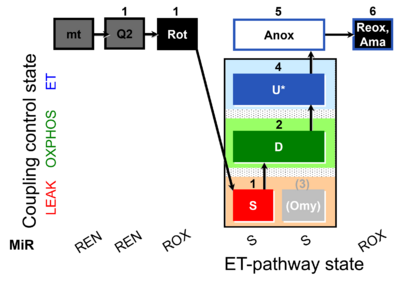 |
| SUIT-007 | Glutamate anaplerosis | 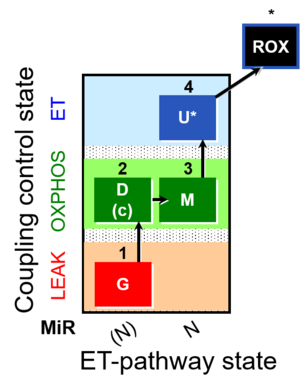 |
| SUIT-007 O2 ce-pce D030 | Glutamate anaplerotic pathway | 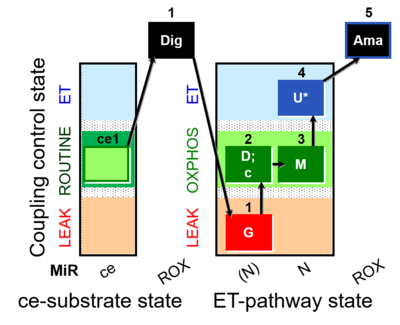 |
| SUIT-008 | PM+G+S_OXPHOS+Rot_ET | 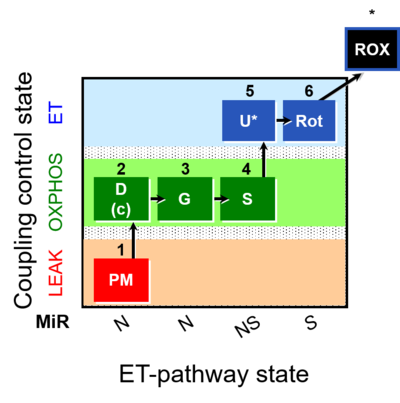 |
| SUIT-008 O2 ce-pce D025 | Q-junction ce-pce | 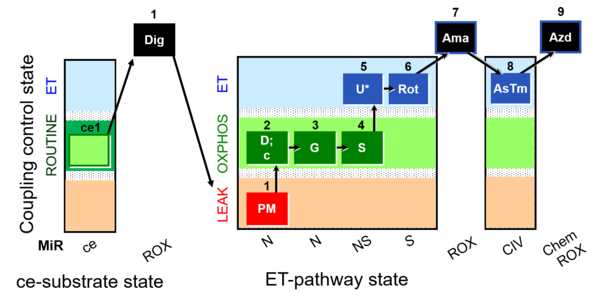 |
| SUIT-008 O2 mt D026 | Q-junction mtprep | 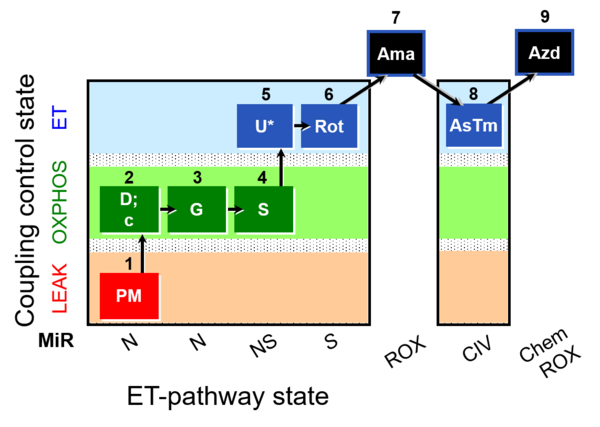 |
| SUIT-008 O2 pce D25 | NS(PGM) | 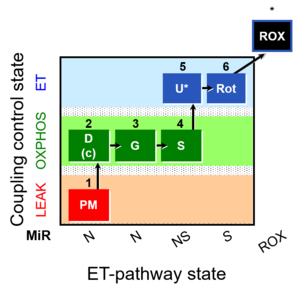 |
| SUIT-009 AmR ce-pce D019 | H2O2 RET ce-pce S_L | 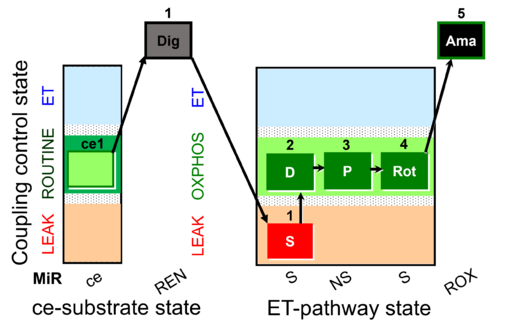 |
| SUIT-009 AmR mt D021 | H2O2 mtprep | 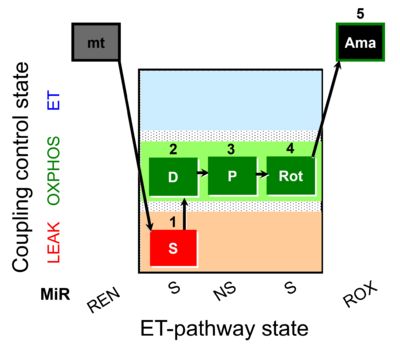 |
| SUIT-010 | Digitonin test | 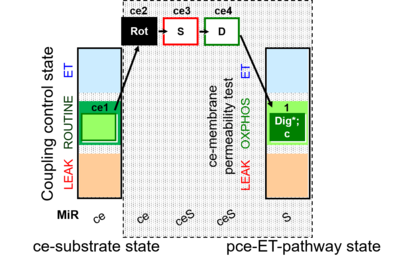 |
| SUIT-010 O2 ce-pce D008 | Dig titration-pce |  |
| SUIT-011 | GM+S_OXPHOS+Rot_ET | 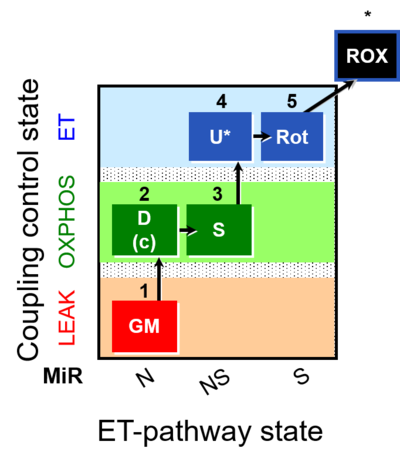 |
| SUIT-011 O2 pfi D024 | NS physiological maximum capapcity in fibres | 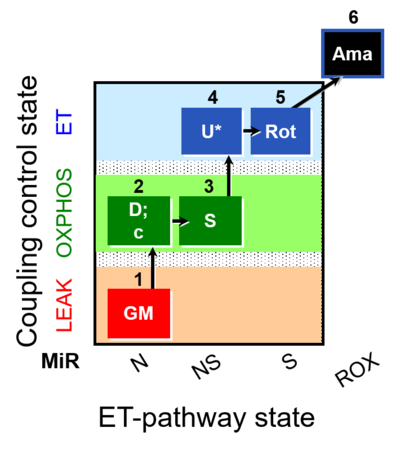 |
| SUIT-012 | PM+G_OXPHOS | 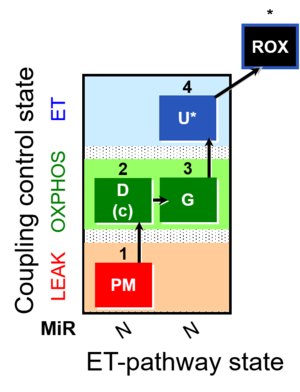 |
| SUIT-012 O2 ce-pce D052 | N(PGM) | 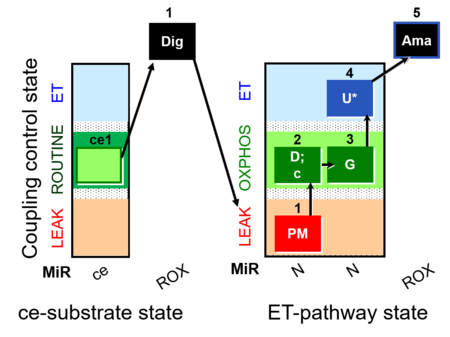 |
| SUIT-012 O2 mt D027 | N CCP mtprep | 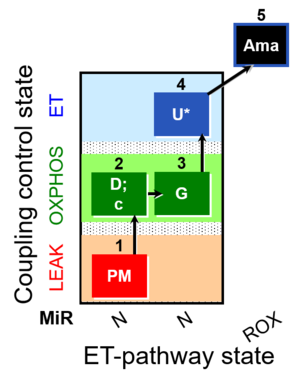 |
| SUIT-013 | ce | 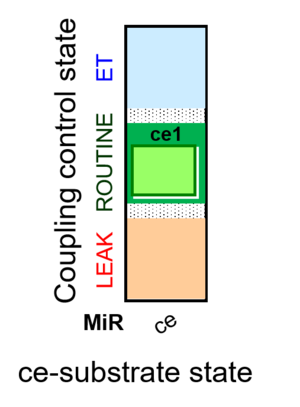 |
| SUIT-013 AmR ce D023 | O2 dependence of H2O2 production ce | 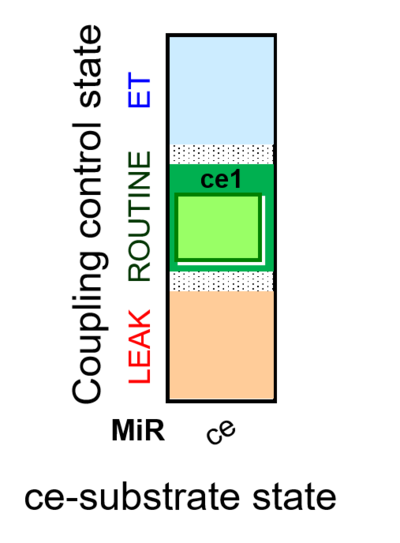 |
| SUIT-014 | GM+P+S_OXPHOS+Rot_ET | 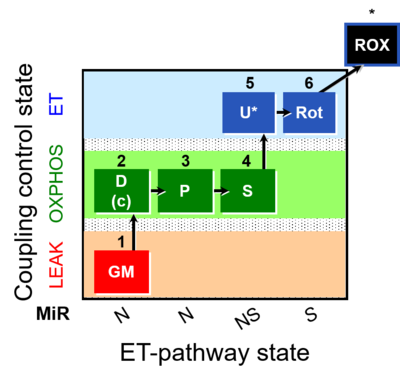 |
| SUIT-014 O2 pfi D042 | NS(PGM) | 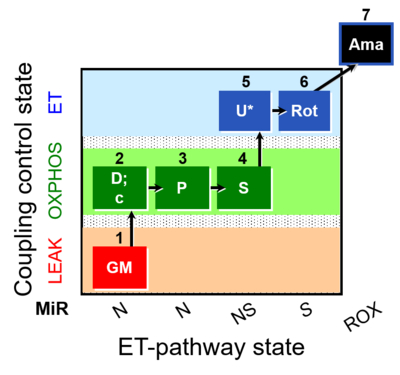 |
| SUIT-015 | F+G+P+S_OXPHOS+Rot_ET | 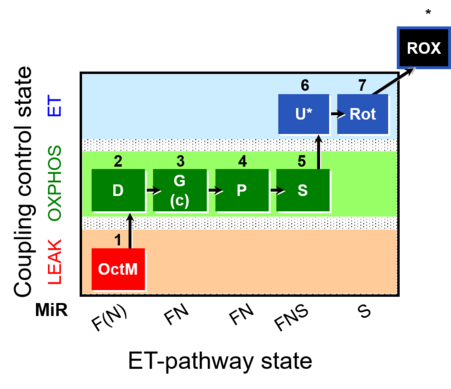 |
| SUIT-015 O2 pti D043 | FNS(Oct,PGM) | 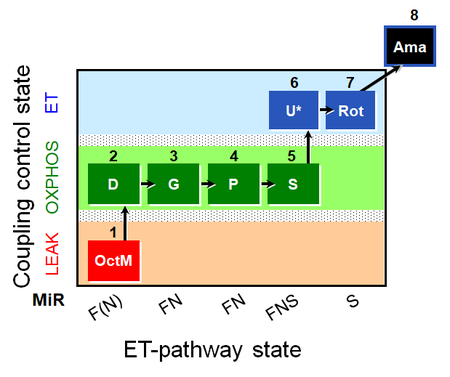 |
| SUIT-016 | F+G+S+Rot_OXPHOS+Omy | 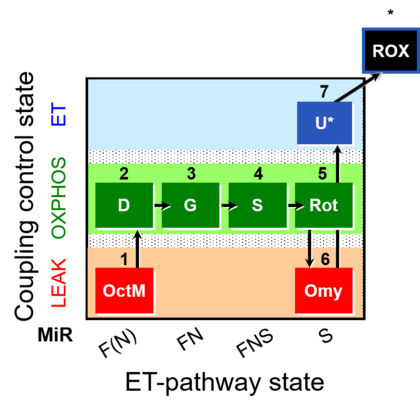 |
| SUIT-016 O2 pfi D044 | FNS(Oct,GM) | 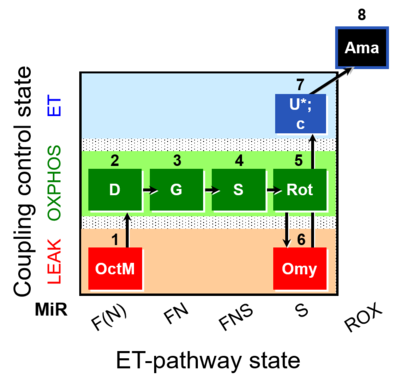 |
| SUIT-017 | F+G+S_OXPHOS+Rot_ET | 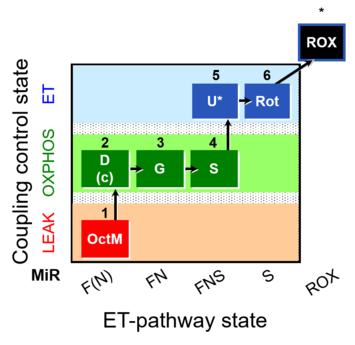 |
| SUIT-017 O2 mt D046 | FNS(Oct,GM) | 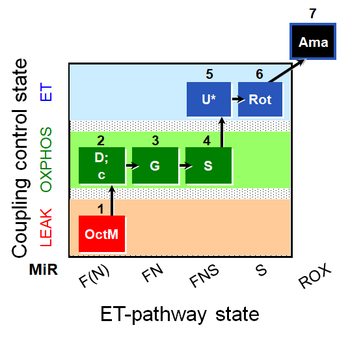 |
| SUIT-017 O2 pfi D049 | FNS(Oct,GM) |  |
| SUIT-018 | O2 dependence-H2O2 | 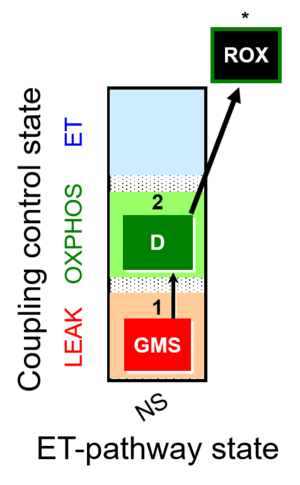 |
| SUIT-018 AmR ce-pce D068 | O2 dependence-H2O2 | 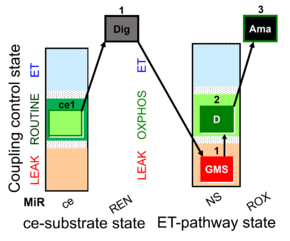 |
| SUIT-018 AmR mt D031 | O2 dependence-H2O2 | 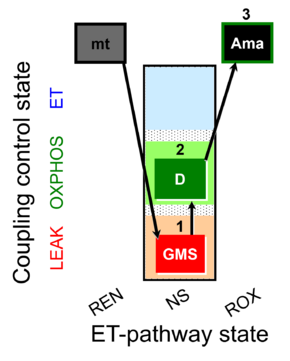 |
| SUIT-018 AmR mt D040 | NS(GM) |  |
| SUIT-018 AmR mt D041 | O2 dependence-H2O2 |  |
| SUIT-019 | Pal+Oct+P+G_OXPHOS+S+Rot_ET | 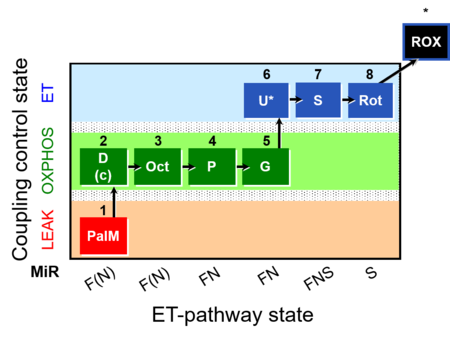 |
| SUIT-019 O2 pfi D045 | FNS(PalOct,PGM) | 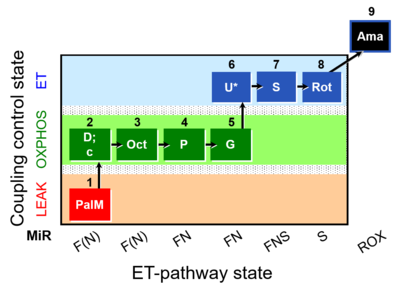 |
| SUIT-020 | PM+G+S+Rot_OXPHOS+Omy | 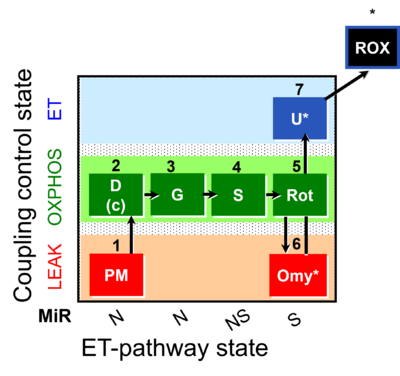 |
| SUIT-020 Fluo mt D033 | NS(PGM) | 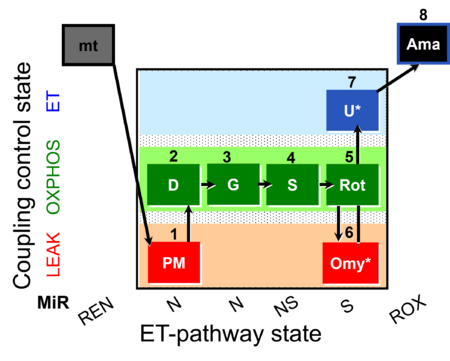 |
| SUIT-020 O2 mt D032 | Q-junction additivity and respiratory control for membrane potential | 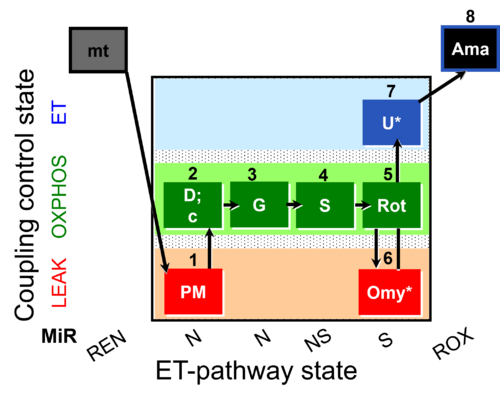 |
| SUIT-021 | OXPHOS (GM+S+Rot+Omy) | 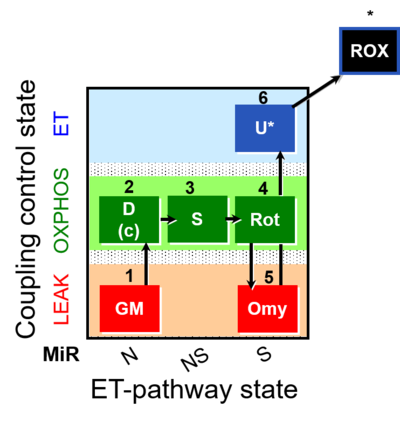 |
| SUIT-021 Fluo mt D036 | NS(GM) | 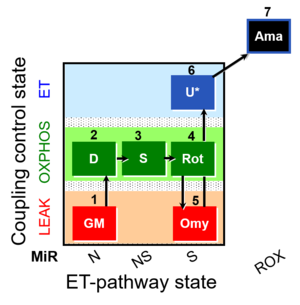 |
| SUIT-021 O2 mt D035 | NS(GM) | 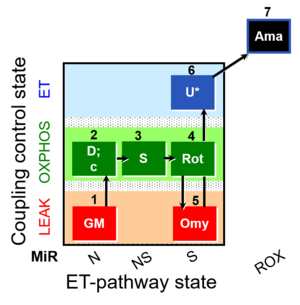 |
| SUIT-022 | AOX (ce CN+SHAM) | 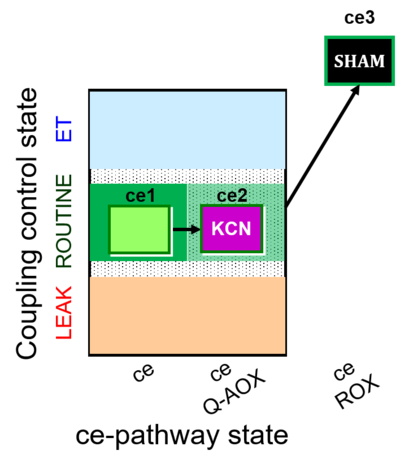 |
| SUIT-022 O2 ce D051 | AOX-ce CN+SHAM | 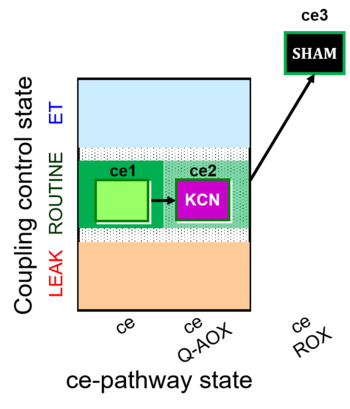 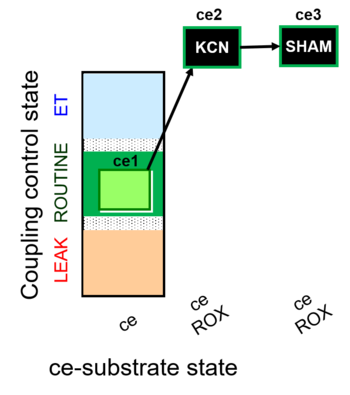 |
| SUIT-023 | AOX-ce SHAM+CN | 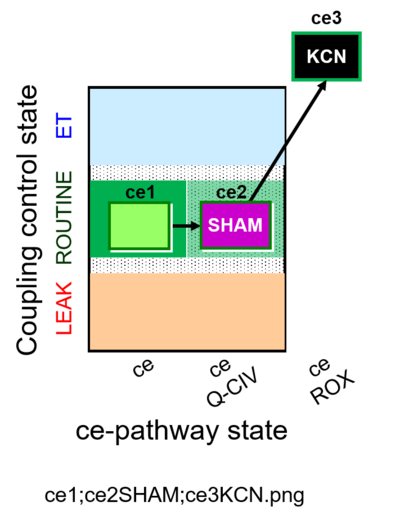 |
| SUIT-023 O2 ce D053 | AOX-ce SHAM+CN | 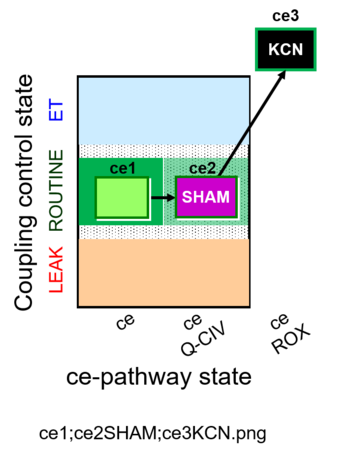 |
| SUIT-024 | ATPase (PM) | 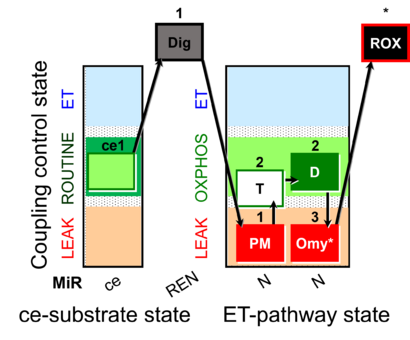 |
| SUIT-024 O2 ce-pce D056 | N(PM) | 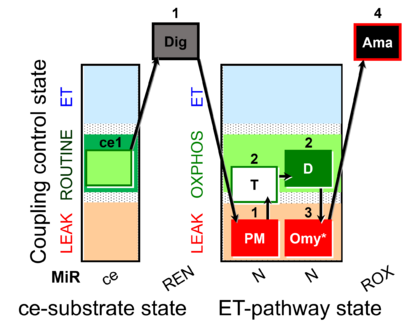 |
| SUIT-025 | OXPHOS (F+M+P+G+S+Rot) | 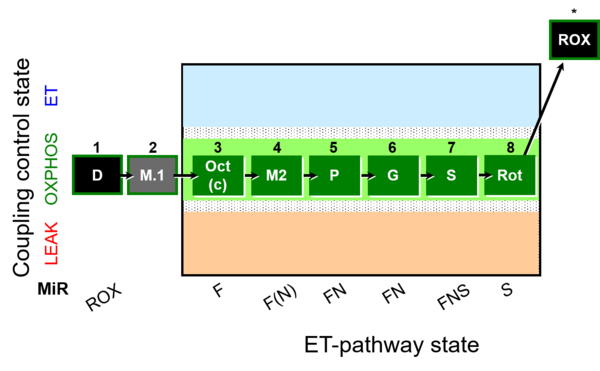 |
| SUIT-025 O2 mt D057 | FNS(Oct,PGM) | 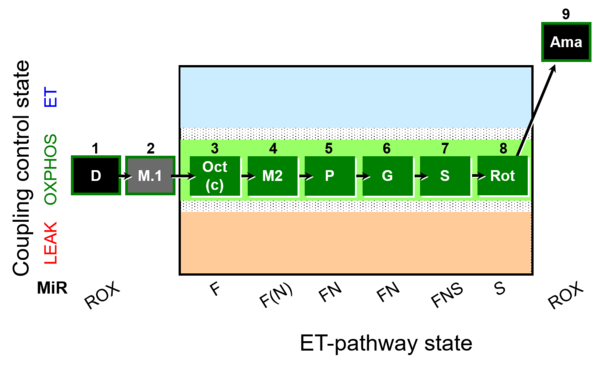 |
| SUIT-026 | RET | 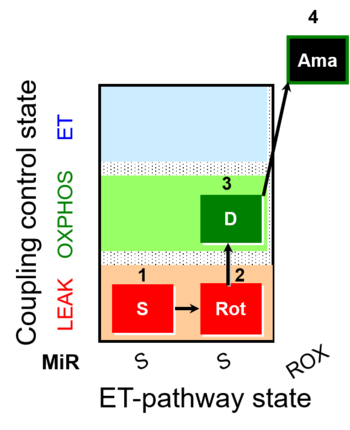 |
| SUIT-026 AmR ce-pce D087 | RET | 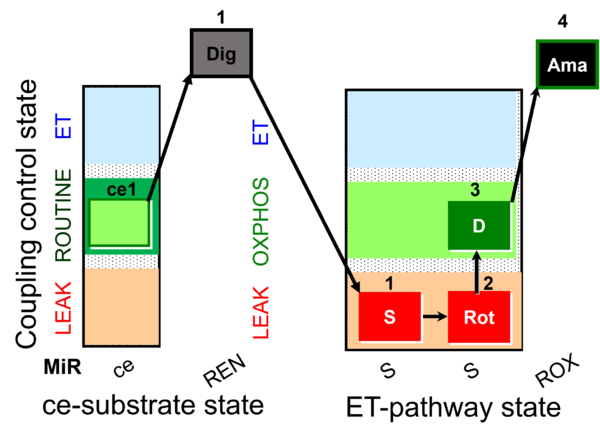 |
| SUIT-026 AmR mt D064 | RET | 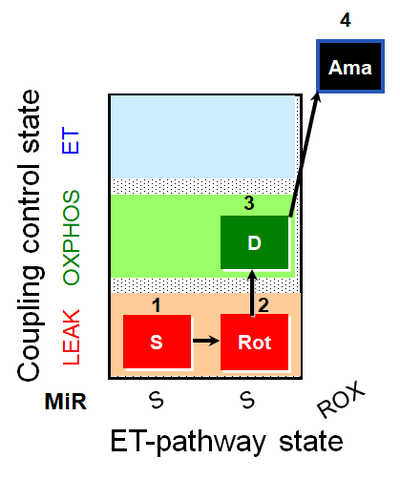 |
| SUIT-026 AmR mt D077 | RET |  |
| SUIT-026 O2 ce-pce D088 | RET (respiratory control) of SUIT-026 AmR ce-pce D087 | 400px |
| SUIT-026 O2 mt D063 | RET (respiratory control) of SUIT-026 AmR mt D064 | 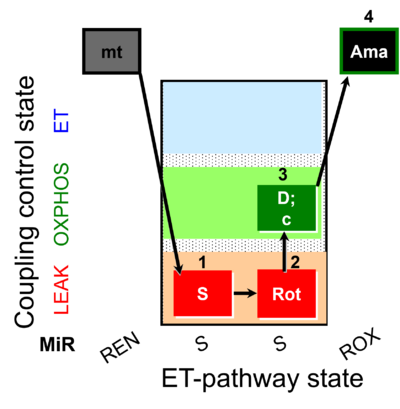 |
| SUIT-027 | Malate anaplerosis | 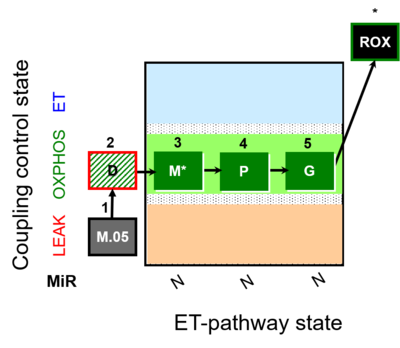 |
| SUIT-027 O2 ce-pce D065 | Malate anaplerosis | 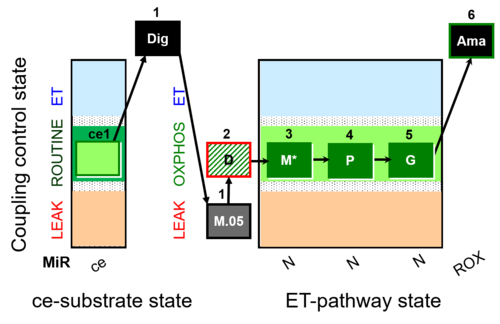 |
| SUIT-028 | NS(PGM) |  |
| SUIT-029 O2 mt D066 | QC_imt_PM_T+OXPHOS+c+Omy_ET_G+S+Rot | 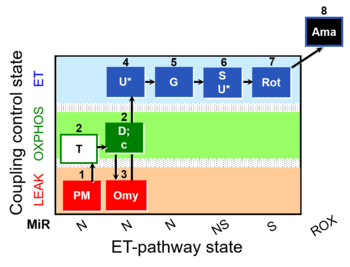 |
| SUIT-031 | PM+S+Rot | 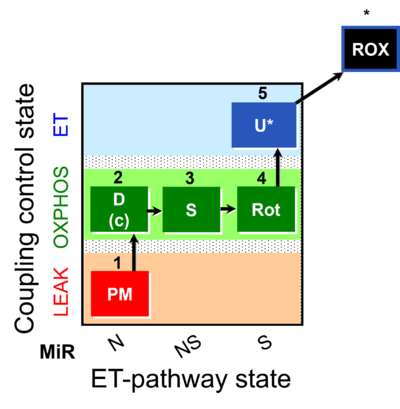 |
| SUIT-031 O2 ce-pce D079 | PM+S+Rot | 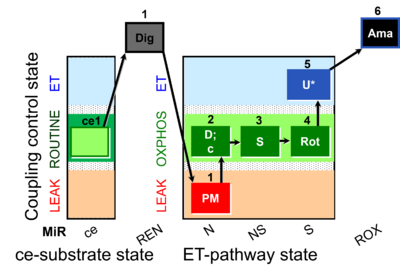 |
| SUIT-031 O2 mt D075 | PM+S+Rot | 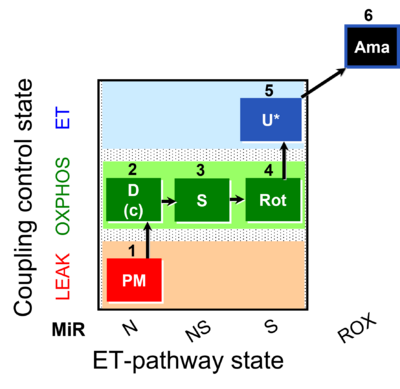 |
| SUIT-031 Q ce-pce D074 | PM+S+Rot | 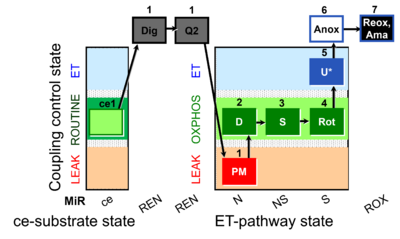 |
| SUIT-031 Q mt D072 | PM+S+Rot | 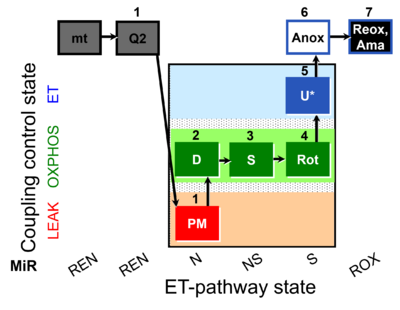 |
| SUIT-033 | CCP-mtprep | 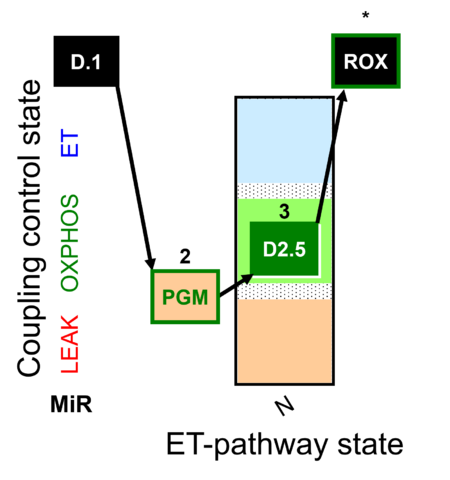 |
| SUIT-036 O2 mt D089 | FAO(Pal) & M kinetics | 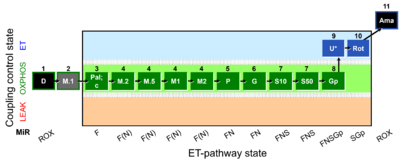 |
| SUIT-037 O2 mt D090 | FAO(Oct) & M kinetics | 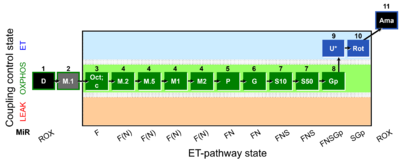 |
| SUIT-038 O2 mt D091 | FAO control & M kinetics | 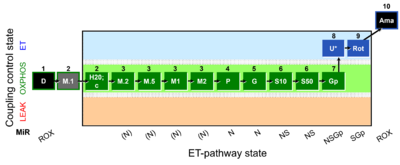 |
| SUIT-039 O2 mt D092 | FAO and NS-pathways | 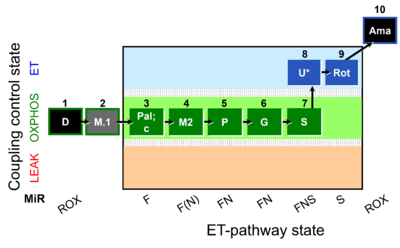 |
| SUIT-039 O2 pfi D093 | FAO and NS-pathways |  |
| SUIT-040 O2 mt D094 | FAO and NSGp-pathways | 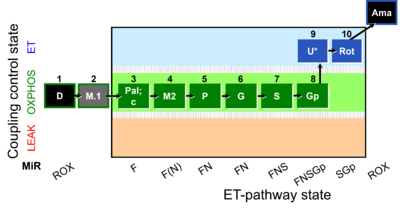 |
| SUIT-040 O2 pfi D095 | FAO and NSGp-pathways |  |
| SUIT-041 O2 mt D096 | Optimum [acylcarnitine] test | 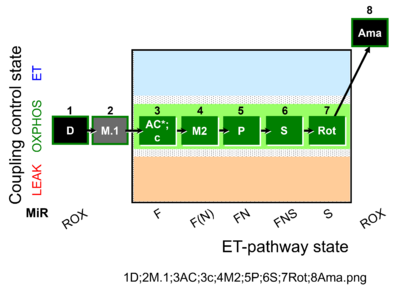 |
| Safranin | Saf | Safranin is one of the most established dyes for measuring mitochondrial membrane potential by fluorometry. It is an extrinsic fluorophore with an excitation wavelength of 495 nm and emission wavelength of 587 nm. Safranin is a potent inhibitor of N-linked respiration and of the phosphorylation system. Synonyms: Safranin O, Safranin Y, Safranin T, Gossypimine, Cotton Red, Basic Red2 |
| Salicylhydroxamic acid | SHAM | Salicylhydroxamic acid (SHAM; synonym: 2-Hydroxybenzohydroxamic acid N,2-Dihydroxybenzamide) is an inhibitor of the alternative oxidase (AOX). When AOX is blocked by SHAM, electrons are forced through the CIII-cytochrome c oxidase pathway, allowing observation of the operation of the CIII-CIV pathway without AOX activity. |
| Sample | s | A sample is one or more parts taken from an ensemble that is studied. A sample is either stored for later quantification or prepared and possibly separated into subsamples, which are enclosed in a system for qualitative or quantitative investigation. A pure sample S is a pure gas, pure liquid or pure solid of a defined elementary entity-type. A pure biological sample is a cell type, tissue, or organism without its solid, liquid or gaseous environment. Then the system used to investigate sample S contains only entities of entity-type S, and the volume VS [L] and mass mS [kg] of the pure (sub)sample S are identical to the volume V and mass m of the experimental system. A pure sample S may be mixed with other components to be investigated as a solution, mixture, or suspension, indicated by the symbol s in contrast to the pure sample S. A sample s is obtained in combination with other components, such that the volume Vs [L] and mass ms [kg] of the sample s are larger than the volume VS and mass mS of the pure sample S. For example, the number of cells Nce [Mx] can be counted in a sample s of a cell suspension, whereas the mass mce [mg] of cells requires a pure sample S of cells to be measured on a mass-balance. Clarity of statistical representation is improved, if the symbol N is used for the number of primary samples taken from a study group, and the symbol n is used for the number of subsamples studied as technical repeats. |
| Sample - DatLab 7 | F3 | In the window Sample, information is entered and displayed for the sample (Sample type, Cohort, Sample code, Sample number, Subsample number and sample concentration). Entries can be edited any time during the experiment in real-time or during post-experiment analysis. All related results are recalculated instantaneously with the new parameters. Initially, the Edit experiment window displays information from the last file recorded and saved while connected to the O2k. |
| Sample and medium - DatLab | F3 | DatLab 8: In the window Sample and medium, information is entered and displayed for the sample and medium. Entries can be edited any time during the experiment or during post-experiment analysis. All related results are recalculated instantaneously with the new parameters. The window can be opened whenever a file is loaded or currently recorded. DatLab 7: Sample |
| Sample mass concentration | Cms | Sample mass concentration is Cms = ms·V-1 [kg·m-3]. |
| Saponin | Sap | Saponin is a mild detergent that permeabilizes plasma membranes completely and selectively due to their high cholesterol content, whereas mt-membranes with lower cholesterol content are affected only at higher concentrations. Applied for permeabilization of muscle fibres. |
| Save - DatLab | Ctrl+S | Save a DatLab file. |
| Save and Disconnect | Ctrl+F4 | Save and Disconnect: Stops data acquisition and disconnects from the O2k (only for DatLab 7) |
| Scaling - DatLab | F6 | Scaling a graph in DatLab provides flexibility to vary the display of the plots and create Graph layouts. It allows viewing a data plot in differently scaled graphs, zooming the signal and time scales, and scrolling along the axes of the graph provide maximum information on the current experiment. This does not influence the format of stored data. Different ranges for the axes change the appearance of data dramatically. It is highly recommended to use reference layouts. »Compare: Select plots - DatLab. |
| Science - the concept | Science | As per the 2017 UNESCO Recommendation on Science and Scientific Researchers, the term ‘science’ signifies the enterprise whereby humankind, acting individually or in small or large groups, makes an organized attempt, in cooperation and in competition, by means of the objective study of observed phenomena and its validation through sharing of findings and data and through peer review, to discover and master the chain of causalities, relations or interactions; brings together in a coordinated form subsystems of knowledge by means of systematic reflection and conceptualization; and thereby furnishes itself with the opportunity of using, to its own advantage, understanding of the processes and phenomena occurring in nature and society. |
| Science Citation Index | SCI | The Science Citation Index SCI offers bibliographical access to a curated collection of journals across 178 scientific disciplines. The SCI provides gold-standard lists of established journals. |
| Second | s | The second, symbol s, is the SI unit of time. It is defined by taking the fixed numerical value of the caesium frequency ∆νCs, the unperturbed ground-state hyperfine transition frequency of the caesium 133 atom, to be 9 192 631 770 when expressed in the unit Hz, which is equal to s−1. |
| Select plots - DatLab | Ctrl+F6 | In the pull-down menue [Graph], Select plots opens the Graph layout window 'Plots'. For each graph, the plots shown with the Y1 or Y2 axis can be selected, axis labels and line styles can be defined, the unit for the calibrated signal can be changed, Flux/Slope can be chosen to be displayed as Flux per volume or as normalized specific flux/flow, the background correction can be switched on or off, and the channel can be selectively displayed as the raw signal. Graph layouts can be selected and loaded or a Graph layout may be saved. »Compare: Scaling - DatLab. |
| Serbian Society for Mitochondrial and Free Radical Physiology | SSMFRP | The Serbian Society for Mitochondrial and Free Radical Physiology (SSMFRP) was established in 2008 as a national Society and has 150 members who gather research in the fields of molecular biology, biochemistry, medicine, chemistry, agriculture, physics and other related disciplines. The SSMFRP was founded as a voluntary non-governmental and non-profit association for researchers whose goal is to support the creative improvement of scientific knowledge about the physiology of mitochondria and free radicals, support for the development of modern research approaches and integration of fundamental research in order to better understand the role of free radicals in pathophysiological states, as well as promoting scientific knowledge in the country and abroad. |
| Set Power O2k number - DatLab | Ctrl+P | DatLab 8: Set a Power-O2k number to label the O2k in a Power O2k-Lab. DatLab 7 : Set the Power-O2k number in the O2k configuration window. |
| Signal-to-noise ratio | S/N | The signal to noise ratio is the ratio of the power of the signal to that of the noise. For example, in fluorimetry it would be the ratio of the square of the fluorescence intensity to the square of the intensity of the background noise. |
| Sirtuins | Sirt | Sirtuins are NAD+-dependent deacetylases which play a prominent role as metabolic regulators. Their dependence on intracellular levels of NAD+ (NAD+ activates sirtuin activity, whereas NADH inhibits it) makes them suitable as sensors that can detect cellular energy status. » MiPNet article |
| Society for Heart and Vascular Metabolism | SHVM | The Society for Heart and Vascular Metabolism (SHVM) The Society for Heart and Vascular Metabolism was founded in 2001, with the intent of providing a forum for the free exchange of ideas by a group of investigators that had a special interest in the multiple roles of intermediary metabolism in the cardiovascular system. An important aim of the Society is to foster interactions between young investigators and senior scientists and our meetings are deliberately designed to maximize these interactions. There is growing recognition across many areas of scientific investigation and in the cardiovascular arena of the importance of metabolic homeostasis. The Society for Heart and Vascular Metabolism intends to remain at the vanguard of this rapidly expanding area. |
| Society for Mitochondrial Research and Medicine - India | SMRM-India | The Society for Mitochondria Research and Medicine - India (SMRM-India) is a nonprofit organization of scientists, clinicians and academicians. The purpose of SMRM is to foster research on basic science of mitochondria, mitochondrial pathogenesis, prevention, diagnosis and treatment through out India and abroad. |
| Sodium fluoride | NaF | Sodium fluoride (NaF) is used in combination with beryllium sulfate to form beryllium trifluoride (BeF3−), to inhibit the ATP synthase if it is exposed by disruption of the mitochondrial membranes. |
| Sodium orthovanadate | VO4 | Sodium vanadate (Na3VO4) is used as an ATPase inhibitor. |
| Sodium phosphate buffer | Na-PB | Sodium phosphate buffer, Na-PB, for HRR with freeze-dried baker´s yeast. |
| Solubility | SG | The solubility of a gas, SG, is defined as concentration divided by partial pressure, SG = cG·pG-1. |
| Speed | v [m·s-1] | Speed, v [m·s-1], is the distance, s [m], covered by a particle per unit time, irrespective of geometrical direction in space. Therefore, speed is not a vector, in contrast to velocity. v = ds/dt [m·s-1] |
| Standard operating procedures | SOP | The following definition is incomplete. Standard operating procedures are a set of step-by-step instructions to achieve a predictable, standardized, desired result often within the context of a longer overall process. |
| State 2 | ROXD |  Substrate limited state of residual oxygen consumption, after addition of ADP to isolated mitochondria suspended in mitochondrial respiration medium in the absence of reduced substrates (ROXD). Residual endogenous substrates are oxidized during a transient stimulation of oxygen flux by ADP. The peak – supported by endogenous substrates – is, therefore, a pre-steady state phenomenon preceding State 2. Subsequently oxygen flux declines to a low level (or zero) at the steady State 2 (Chance and Williams 1955). ADP concentration (D) remains high during ROXD. Substrate limited state of residual oxygen consumption, after addition of ADP to isolated mitochondria suspended in mitochondrial respiration medium in the absence of reduced substrates (ROXD). Residual endogenous substrates are oxidized during a transient stimulation of oxygen flux by ADP. The peak – supported by endogenous substrates – is, therefore, a pre-steady state phenomenon preceding State 2. Subsequently oxygen flux declines to a low level (or zero) at the steady State 2 (Chance and Williams 1955). ADP concentration (D) remains high during ROXD. |
| State 3 | P | |
| State 3u | E | |
| State 4 | LT | |
| Static head | L |
In a completely coupled system, not only the output flux but also the input flux are zero at static head, which then is a state of ergodynamic equilibrium (Gnaiger 1993b). Whereas the output force is maximum at ergodynamic equilibrium compensating for any given input force, all forces are zero at thermodynamic equilibrium. Flows are zero at both types of equilibria, hence entropy production or power (power = flow x force) are zero in both cases, i.e. at thermodynamic equilibrium in general, and at ergodynamic equilibrium of a completely coupled system at static head. |
| Statistical significance | p | It is advisable to replace levels of statistical significance (*, **, ***) by simply stating the actual p-values. |
| Stirrer A on/off | F11 | The stirrer in chamber A is switched on/off. |
| Stirrer B on/off | F12 | The stirrer in chamber B is switched on/off. |
| Stirrer power | F11, F12 | Stirrer power is switched on and off during operation of the Oroboros O2k in DatLab by pressing [F11] (left chamber) and [F12] (right chamber), respectively. This is functional only with a stirrer bar added to each O2k chamber. |
| Stoichiometric number | νX | The sign of the stoichiometric number νX is determined by the nonspatial direction of the transformation (positive for products, negative for substrates), and the magnitude of νX is determined by the stoichiometric form. For instance, νA=-1 in the reaction 0 = -1 A + 2 B (-1 glucose converted to +2 lactate), but νA=-1/6 in the reaction 0 = -1/6 A - 1 B + 1 C (-1/6 glucose and -1 O2 converted to +1 H2CO3). |
| Submitochondrial particles | smtp | Submitochondrial particles (smtp) consist of membrane fragments which retain most of the enzymatic machinery required in electron transfer and oxidative phosphorylation. Such membrane fragments are continuous closed vesicles formed by resealing of mt-membrane fragments after disruption of the mitochondrial structure. smtp are used to isolate the inner-membrane-bound ET pathway (mETS) from the upstream modules of the Electron transfer pathway (ETS) which are located in the mt-matrix and outer mt-membrane (transporters). smtp are obtained by treatment of mitochondria with membrane-dispersing agents such as digitonin at high concentration or by sonic irradiation. |
| Subsample | n | Subsamples can be obtained (1) from a homogenous sample (e.g. cell suspension, tissue homogenate, isolated mitochondria), (2) as subsamples obtained by splitting a sample into comparable parts (e.g. permeabilized muscle fibres from a biopsy split into different chambers for repeated measurements), or (3) repetitive sampling (e.g. taking multiple biopsies) at a single time point. Subsamples may be used for (i) application of different types of assay (e.g. for measurement of respiration and enzyme activities), and (ii) a number of repetitions, n, of the same assay on the same sample. |
| Substrate-uncoupler-inhibitor titration | SUIT | Mitochondrial Substrate-uncoupler-inhibitor titration (SUIT) protocols are used with mitochondrial preparations to study respiratory control in a sequence of coupling and substrates states induced by multiple titrations within a single experimental assay. |
| Substrates as electron donors | Sred | Substrates as electron donors are reduced fuel compounds Sred that are oxidized to an oxidized product Pox during H+-linked electron transfer, Sred → Pox + 2{H+ + e-}. Mitochondrial respiration depends on a continuous flow of electron-supplying substrates across the mitochondrial membranes into the matrix space. Many substrates are strong anions that cannot permeate lipid membranes and hence require carriers. |
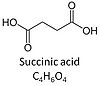
| Succinate | S |
Succinic acid, C4H6O4, (butanedioic acid) is a dicarboxylic acid which occurs under physiological conditions as the anion succinate2-, S, with pKa1 = 4.2 and pKa2 = 5.6. Succinate is formed in the TCA cycle, and is a substrate of CII, reacting to fumarate and feeding electrons into the Q-junction. Succinate (CII-linked) and NADH (CI-linked) provide convergent electron entries into the Q-junction. Succinate is transported across the inner mt-membrane by the dicarboxylate carrier. The plasma membrane of many cell types is impermeable for succinate (but see Zhunussova 2015 Am J Cancer Res for an exception). Incubation of mt-preparations by succinate alone may lead to accumulation of oxaloacetate, which is a potent inhibitor of Complex II (compare Succinate and rotenone). High activities of mt-Malic enzyme (mtME) prevent accumulation of oxaloacetate in incubations with succinate without rotenone. |
| Succinate dehydrogenase | SDH | Succinate dehydrogenase is a TCA cycle enzyme converting succinate to fumarate while reducing FAD to FADH2. SDH is the largest component of the mt-inner membrane Complex II (CII) and thus part of the TCA cycle and electron transfer pathway. |
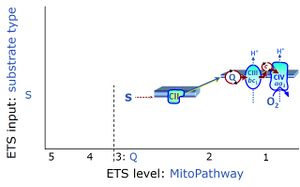
| Succinate pathway | S, SRot |
The Succinate pathway (S-pathway; S) is the electron transfer pathway that supports succinate-linked respiration (succinate-induced respiratory state; previously used nomenclature: CII-linked respiration; SRot; see Gnaiger 2009 Int J Biochem Cell Biol). The S-pathway describes the electron flux through Complex II (CII; see succinate dehydrogenase, SDH) from succinate and FAD to fumarate and CII-bound flavin adenine dinucleotide (FADH2) to the Q-junction. The S-pathway control state is usually induced in mt-preparations by addition of succinate&rotenone. In this case, only Complex III and Complex IV are involved in pumping protons from the matrix (positive phase, P-phase) to the negative phase (N-phase) with a P»/O2 of 3.5 (P»/O ratio = 1.75). |
| Succinyl-CoA ligase | SUCLA, SUCLG | Succinyl-CoA ligase (SUCLA or SUCLG) is a TCA cycle enzyme converting succinyl-CoA + ADP or (GDP) + Pi to succinate + ATP (GTP). Two different isoforms exsist: SUCLA (EC: 6.2.1.5) is the ATP-forming isoenzyme, SUCLG (EC: 6.2.1.4) is the GTP-forming isoenzyme. Both reactions are reversible. This reaction is attributed to mitochondrial substrate-level phosphorylation, which is considered as an alternative way of ATP synthesis because it is partially independent from the respiratory chain and from the mitochondrial proton motive force. |
| Sulfide quinone reductase | SQR | Sulfide quinone reductase (SQR) is involved in electron transfer from sulfide which is used as a hydrogen donor by the mitochondrial respiratory system. SQR is associated with a dioxygenase and a sulfur transferase to release thiosulfate (H2S2O3). |
| Sulfite oxidase | SO | Sulfite oxidase (SO) is a dimeric enzyme, located in the intermembrane space of mitochondria, with each monomer containing a single Mo cofactor and cyt b5-type heme [1]. SO catalyzes the oxidation of sulfite to sulfate as the terminal step in the metabolism of sulfur amino acids and is vital for human health. Inherited mutations in SO result in severe neurological problems, stunted brain growth, and early death [2]. Function: SO catalyzes the terminal reaction in the oxidative degradation of sulfur amino acids with the formation of a sulfate, electrons pass to cytochrom c and are further utilized in the respiratory system. Sulfite + O2 + H2O --> Sulfate + H2O2 Localization: The level of expression of SO differs in various tissues with main predominant localization in liver, kidney, skeletal muscle, heart, placenta, and brain in humans and liver, kidney, heart, brain, and lung in rats [3]. Deficiency: SO is vital for metabolic pathways of sulfur amino acids (cysteine and methionine). Complete lack of this enzyme, typically caused by gene mutation, leads to lethal disease called sulfite oxidase deficiency characterized by neurological abnormalities with brain atrophy. |
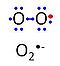
| Superoxide | O2•- |
Superoxide anion, O2•-, is a free radical formed in a one-electron reduction of molecular oxygen (red bullet in the figure), yielding a negatively charged molecule with a single unpaired electron (blue bullet on the left). It is highly reactive with organic compounds, and its intracellular concentration is kept under control by superoxide dismutase. |
| Superoxide dismutase | SOD | Mammalian superoxide dismutase (SOD) exists in three forms, of which the Mn-SOD occurs in mitochondria (mtSOD, SOD2; 93 kD homotetramer) and many bacteria, in contrast to the Cu-Zn forms of SOD (cytosolic SOD1, extracellular SOD3 anchored to the extracellular matrix and cell surface). Superoxide anion (O2•-) is a major reactive oxygen species (ROS) which is dismutated by SOD to oxygen and H2O2. |
| TIP2k - DatLab | F8 | The Titration-Injection microPump (TIP2k) provides automated injection of liquids into both O2k chambers. It is controlled via DatLab, allowing for programmable titration regimes and feedback control. |
| TMPD | Tm | N,N,N',N'-Tetramethyl-p-phenylenediamine dihydrochloride, TMPD, is applied as an artificial substrate for reducing cytochrome c in the respirometric assay for cytochrome c oxidase (CIV) activity. It is maintained in a reduced state by ascorbate and undergoes autoxidation as a function of oxygen pressure, TMPD, ascorbate and cytochrome c concentration. |
| TMRM | TMRM | TMRM (tetramethylrhodamine methyl ester) is an extrinsic fluorophore used as a probe to determine changes in mitochondrial membrane potential. TMRM is a lipophilic cation that is accumulated in the mitochondrial matrix in proportion to Δψmt. Upon accumulation of the dye it exhibits a red shift in its absorption and fluorescence emission spectrum. The fluorescence intensity is quenched when the dye is accumulated in the mitochondrial matrix. |
| Taiwan Society for Mitochondrial Research and Medicine | TSMRM | The Taiwan Society for Mitochondrial Research and Medicine (TSMRM) is a member of ASMRM. |
| Tetrachloro-2-trifluoromethylbenzimidazole | TTFB | 4,5,6,7-Tetrachloro-2-trifluoromethylbenzimidazole is a protonophore or uncoupler of oxidative phosphorylation. |
| Tetrahydrofolate | THF | Tetrahydrofolate, THF, is the substrate in mitochondrial folate-mediated 1C metabolism, an NADH-linked pathway leading to the formation of formate which is exported to the cytosol. |
| Tetraphenylphosphonium | TPP+ | Tetraphenylphosphonium (TPP+). A lipophilic molecular probe in conjunction with an ion selective electrode (ISE) for measuring the mitochondrial membrane potential. |
| The North American Mitochondrial Disease Consortium | NAMDC |
The North American Mitochondrial Disease Consortium (NAMDC) was established to create a network of all clinicians and clinical investigators in North America (US and Canada, with the hope of including Mexico in the future) who follow sizeable numbers of patients with mitochondrial diseases and are involved or interested in mitochondrial research. The NAMDC has created a clinical registry for patients, in the hopes of standardizing diagnostic criteria, collecting important standardized information on patients, and facilitating the participation of patients in research on mitochondrial diseases. For the study of any rare disease, the collection of specimens is a major challenge. The NAMDC is establishing a repository for specimens and DNA from patients with mitochondrial diseases, in order to make materials easily available to consortium researchers. Finally, the NAMDC will conduct clinical trials and other kinds of research. The consortium makes biostatisticians, data management experts, and specialists in clinical research available to participating physicians, so that experiments conducted through the NAMDC can make the most efficient and innovative use of the generous participation of patients. |
| Thenoyltrifluoroacetone | TTFA | Thenoyltrifluoroacetone TTFA is a noncompetitive inhibitor of CII binding on the quinone-binding (SDHC/SDHD). |
| Thioredoxin reductase | TrxR | Thioredoxin reductase (TrxR) is a family of enzymes able to reduce thioredoxin in mammals. |
| Tissue homogenate | thom | A tissue homogenate (thom) is obtained through mechanical micro-disruption of fresh tissue and the cell membranes are mechanically permeabilized. |
| Transmittance | T | When light enter a sample, transmittance (T) is the fraction of the intensity (I) of the light emerging from the sample compared with the incident light intensity (I0): T = I/I0. |
| Tricarboxylic acid cycle | TCA cycle | The tricarboxylic acid (TCA) cycle is a system of enzymes in the mitochondrial matrix arranged in a cyclic metabolic structure, including dehydrogenases that converge in the NADH pool and succinate dehydrogenase (on the inner side of the inner mt-membrane) for entry into the membrane-bound ET pathway mET pathway. Citrate synthase is a marker enzyme of the TCA cycle, at the gateway into the cycle from pyruvate via acetyl-CoA. It is thus the major module of the Electron transfer pathway, upstream of the inner Membrane-bound ET pathway (mET-pathway) and downstream of the outer mt-membrane. Sections of TCA cycle are required for fatty acid oxidation (FAO, β-oxidation). Anaplerotic reactions fuel the TCA cycle with other intermediary metabolites. In the cell, the TCA cycle serves also biosynthetic functions by metabolite export from the matrix into the cytosol. |
| Triethyltin bromide | TET | Triethyltin bromide (TET) is a lipophilic [1] inhibitor of the mitochondrial ATP synthase [2] which is used to induce LEAK state in living cells of Saccharomyces cerevisiae. |
| Uncoupler | U | An uncoupler is a protonophore (CCCP, FCCP, DNP, SF6847) which cycles across the inner mt-membrane with transport of protons and dissipation of the electrochemical proton gradient. Mild uncoupling may be induced at low uncoupler concentrations, the noncoupled state of ET capacity is obtained at optimum uncoupler concentration for maximum flux, whereas at higher concentrations an uncoupler-induced inhibition is observed. |
| Uncoupling protein 1 | UCP1 | Uncoupling protein 1 (UCP1) is also called thermogenin and is predominantly found in brown adipose tissue (BAT). UCP1 belongs to the gene family of uncoupling proteins. It is vital for the maintenance of body temperature, especially for small mammals. As the essential component of non-shivering thermogenesis, it possesses the ability to build and open a pore in the inner mitochondrial membrane through which protons may flow along their electrochemical gradient, generated by respiration, bypassing the ATP-producing re-entry site at the F1F0-ATP synthase. Thereby the energy stored in the electrochemical gradient is dissipated as heat. |
| Uncoupling protein 2 | UCP2 | Uncoupling protein 2 (UCP2) belongs to the gene family of uncoupling proteins. Whereas UCP1 acts as an uncoupler, this may not be the case for UCP2. |
| Uncoupling proteins | UCP | Uncoupling proteins (UCPs) are mitochondrial anion carrier proteins that can be found in the inner mitochondrial membranes of animals and plants. UCP1 acts as an uncoupler by dissipating the electrochemical proton gradient (mitochondrial membrane potential), generated by the electron transfer pathway by pumping protons from the mitochondrial matrix to the mitochondrial intermembrane space. |
| Uncoupling-control ratio | UCR | The uncoupling-control ratio UCR is the ratio of ET-pathway/ROUTINE-respiration (E/R) in living cells, evaluated by careful uncoupler titrations (Steinlechner et al 1996). Compare ROUTINE-control ratio (R/E) (Gnaiger 2008). |
| Uninterrupted power supply | UPS | A back-up power supply may be required to secure uninterrupted power supply. |
| Unit | UX [x]; uQ |
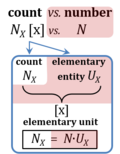
A unit is defined as 'a single individual thing' in Euclid's Elements (Book VII). This defines the elementary entity UX of entity-type X (thing). The International System of Units defines the unit as 'simply a particular example of the quantity concerned which is used as a reference'. Then the value of a quantity Q is the product of a number N and a unit uQ. The symbols UX and uQ are chosen here with U and u for 'unit': UX is the Euclidean or entetic unit ('eunit'), and uQ is the abstract unit ('aunit'). Subscripts X and Q for 'entity-type' and 'quantity-type' reflect perhaps even more clearly than words the contrasting meanings of the two fundamental definitions of an entetic versus abstract 'unit'. The term 'unit' with its dual meanings is used and confused in practical language and the scientific literature today. In the elementary entity UX, the unit (the 'one') relates to the entity-type X, to the single individual thing (single individual or undivided; the root of the word thing has the meaning of 'assembly'). The quantity involved in the unit of a single thing is the count, NX = N·UX [x]. In contrast to counting, a unit uQ is linked to the measurement of quantities Qu = N·uQ, such as volume, mass, energy; and these quantities — and hence the units uQ — are abstracted from entity-types, pulled away from the world of real things. The new SI (2019-05-20) has completed this total abstraction of units, from the previous necessity to not only provide a quantitative definition but also a physical realization of a unit in the form of an 'artefact', such as the international prototype (IPK) for the unit kilogram. The new definitions of the base SI units are independent of any physical realization: uQ is separate from X. The classical unit of Euklid is the elementary unit for counting, entirely independent of measuring. Therefore, the quantity count is unique with respect to two properties: (1) in contrast to all other quantities in the metric system, the count depends on quantization of entities X; and (2) in the SI, the Number 1 is the Unit of the Count of entities — NUCE. |
| United Mitochondrial Disease Foundation | UMDF |
The United Mitochondrial Disease Foundation (UMDF) was founded in 1996 to promote research and education for the diagnosis, treatment and cure of mitochondrial disorders and to provide support to affected individuals and families. |
| VO2max | VO2max; VO2max/M | Maximum oxygen consumption, VO2max, is and index of cardiorespiratory fitness, measured by spiroergometry on human and animal organisms capable of controlled physical exercise performance on a treadmill or cycle ergometer. VO2max is the maximum respiration of an organism, expressed as the volume of O2 at STPD consumed per unit of time per individual object [mL.min-1.x-1]. If normalized per body mass of the individual object, M [kg.x-1], mass specific maximum oxygen consumption, VO2max/M, is expressed in units [mL.min-1.kg-1]. |
| Velocity | v [m·s-1] | Velocity, v [m·s-1], is the speed in a defined spatial direction, and as such velocity is a vector. Velocity is the advancement in distance per unit time, v ≡ dz ∙ dt-1 [m·s-1] |
| Viable cells | vce | Viable cells vce are characterized by an intact plasma membrane barrier function. The total cell count (Nce) is the sum of viable cells (Nvce) and dead cells (Ndce). |
| Viruses and mitochondrial medicine | Virus-mt-Medicine | Not enough is known about viruses and mitochondrial medicine, although several studies point towards a link between viral infection and mitochondrial dysfunction using high-resolution respirometry, with potential impact on drug development. |
| Viton | n.a. | Viton® is a fluoroelastomer with excellent resistance to aggressive fuels and chemicals. Viton is resistant against oxygen diffusion which makes it an ideal material for high-resolution respirometry (Viton O-rings). |
| Volume | V [m3]; 1 m3 = 1000 L | Volume V is a derived quantity based on the SI base quantity length [m] and is expressed in terms of SI base units in the derived unit cubic meter [m3]. The liter [L = dm3] is a conventional unit of volume for concentration and is used for most solution chemical kinetics. The volume V contained in a system (experimental chamber) is separated from the environment by the system boundaries; this is called the volume of the system, and described in practical language as big/small (derived from length, height) or voluminous. Systems are defined at constant volume or constant pressure. For a pure sample S, the volume VS of the pure sample equals the volume V of the system, VS = V. For sample s in a mixture, the ratio Vs·V-1 is the nondimensional volume fraction Φs of sample s. Quantities divided by volume are concentrations of sample s in a mixture, such as count concentration CX = NX·V-1 [x·L-1], and amount of substance concentration CB = nB·V-1 [mol·L-1]. Mass concentration is density ρs = ms·V-1 [kg·L-1]. In closed compressible systems (with a gas phase), the concentration of the gas increases, when pressure-volume work is performed on the system. |
| Water | H2O |
Water, H2O, is widely used in the laboratory, particularly as a solvent and cleaning agent. Chemically pure water is prepared in various grades of purification: double distilled water (ddH2O) versus distilled water (dH2O or aqua destillata, a.d.) and deionized or demineralized water (diH2O) with various combination purification methods. When H2O is mentioned without further specification in published protocols, it is frequently assumed that the standards of each laboratory are applied as to the quality of purified water. Purification is not only to be controlled with respect to salt content and corresponding electrical conductivity (ultra-pure water: 5.5 μS/m due to H+ and OH- ions), but also in terms of microbial contamination. |
| Wet mass | Mw | The wet mass of a tissue or biological sample, obtained after blotting the sample to remove an arbitrary amount of water adhering externally to the sample. |
| Work | deW [J] | Work [J] is a specific form of energy in the First Law of thermodynamics, and a specific form of exergy in the Second Law of thermodynamics, performed by a closed or open system on its surroundings (the environment). This is the definition of external work, which is zero in isolated systems. The term exergy includes external and internal work. Mechanical work is force [N] times path length [m]. The internal-energy change of a closed system, dU, is due to external exchange (e) of work and heat, and external total work (et, including pressure-volume work) is the internal-energy change minus heat, detW = dU - deQ |
| Zero calibration | R0 | Zero calibration is, together with air calibration, one of the two steps of the POS calibration. It is performed in the closed chamber after all the oxygen has been depleted by the addition of dithionite or by respiration of imt or cells. Any incubation medium can be used for zero calibration with dithionite or sample. Unlike air calibration, it is not necessary to perform a zero calibration on each experimental day. After performing a zero calibration, it is recommended not running other experiments on the same day. Even after standard cleaning of the O2k-chambers, there might be residual amounts of reduced dithionite in the chamber, affecting the oxygen flux in subsequent experiments performed on the same day. |
| ≡ | ≡ | The symbol ≡ indicates (numerical) equivalence, in contrast to = as the symbol for (physicochemical) equality. |